A microfluidics-based in vitro model of the gastrointestinal human-microbe interface
- PMID: 27168102
- PMCID: PMC4865890
- DOI: 10.1038/ncomms11535
A microfluidics-based in vitro model of the gastrointestinal human-microbe interface
Abstract
Changes in the human gastrointestinal microbiome are associated with several diseases. To infer causality, experiments in representative models are essential, but widely used animal models exhibit limitations. Here we present a modular, microfluidics-based model (HuMiX, human-microbial crosstalk), which allows co-culture of human and microbial cells under conditions representative of the gastrointestinal human-microbe interface. We demonstrate the ability of HuMiX to recapitulate in vivo transcriptional, metabolic and immunological responses in human intestinal epithelial cells following their co-culture with the commensal Lactobacillus rhamnosus GG (LGG) grown under anaerobic conditions. In addition, we show that the co-culture of human epithelial cells with the obligate anaerobe Bacteroides caccae and LGG results in a transcriptional response, which is distinct from that of a co-culture solely comprising LGG. HuMiX facilitates investigations of host-microbe molecular interactions and provides insights into a range of fundamental research questions linking the gastrointestinal microbiome to human health and disease.
Conflict of interest statement
The authors have a corresponding patent application (WO/2013/144253), which is currently pending.
Figures

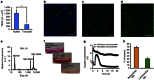
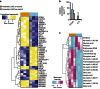

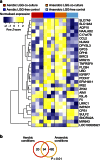

References
-
- Gerber G. K. The dynamic microbiome. FEBS Lett. 588, 4131–4139 (2014). - PubMed
-
- Knip M. & Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 12, 154–167 (2016). - PubMed
-
- Zhao L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 11, 639–647 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

