TAC102 Is a Novel Component of the Mitochondrial Genome Segregation Machinery in Trypanosomes
- PMID: 27168148
- PMCID: PMC4864229
- DOI: 10.1371/journal.ppat.1005586
TAC102 Is a Novel Component of the Mitochondrial Genome Segregation Machinery in Trypanosomes
Erratum in
-
Correction: TAC102 Is a Novel Component of the Mitochondrial Genome Segregation Machinery in Trypanosomes.PLoS Pathog. 2016 Jul 1;12(7):e1005750. doi: 10.1371/journal.ppat.1005750. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27367146 Free PMC article.
Abstract
Trypanosomes show an intriguing organization of their mitochondrial DNA into a catenated network, the kinetoplast DNA (kDNA). While more than 30 proteins involved in kDNA replication have been described, only few components of kDNA segregation machinery are currently known. Electron microscopy studies identified a high-order structure, the tripartite attachment complex (TAC), linking the basal body of the flagellum via the mitochondrial membranes to the kDNA. Here we describe TAC102, a novel core component of the TAC, which is essential for proper kDNA segregation during cell division. Loss of TAC102 leads to mitochondrial genome missegregation but has no impact on proper organelle biogenesis and segregation. The protein is present throughout the cell cycle and is assembled into the newly developing TAC only after the pro-basal body has matured indicating a hierarchy in the assembly process. Furthermore, we provide evidence that the TAC is replicated de novo rather than using a semi-conservative mechanism. Lastly, we demonstrate that TAC102 lacks an N-terminal mitochondrial targeting sequence and requires sequences in the C-terminal part of the protein for its proper localization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
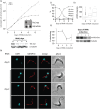
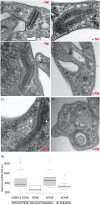


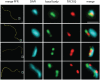
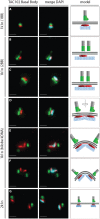


Comment in
-
A novel component of the mitochondrial genome segregation machinery in trypanosomes.Microb Cell. 2016 Jul 28;3(8):352-354. doi: 10.15698/mic2016.08.519. Microb Cell. 2016. PMID: 28357371 Free PMC article.
Similar articles
-
Characterization of the novel mitochondrial genome segregation factor TAP110 in Trypanosoma brucei.J Cell Sci. 2021 Mar 8;134(5):jcs254300. doi: 10.1242/jcs.254300. J Cell Sci. 2021. PMID: 33589495 Free PMC article.
-
p166 links membrane and intramitochondrial modules of the trypanosomal tripartite attachment complex.PLoS Pathog. 2022 Jun 16;18(6):e1010207. doi: 10.1371/journal.ppat.1010207. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35709300 Free PMC article.
-
Molecular model of the mitochondrial genome segregation machinery in Trypanosoma brucei.Proc Natl Acad Sci U S A. 2018 Feb 20;115(8):E1809-E1818. doi: 10.1073/pnas.1716582115. Epub 2018 Feb 6. Proc Natl Acad Sci U S A. 2018. PMID: 29434039 Free PMC article.
-
Failure is not an option - mitochondrial genome segregation in trypanosomes.J Cell Sci. 2018 Sep 17;131(18):jcs221820. doi: 10.1242/jcs.221820. J Cell Sci. 2018. PMID: 30224426 Review.
-
Beyond replication: division and segregation of mitochondrial DNA in kinetoplastids.Mol Biochem Parasitol. 2014 Aug;196(1):53-60. doi: 10.1016/j.molbiopara.2014.03.008. Epub 2014 Apr 4. Mol Biochem Parasitol. 2014. PMID: 24704441 Review.
Cited by
-
A fixable probe for visualizing flagella and plasma membranes of the African trypanosome.PLoS One. 2018 May 16;13(5):e0197541. doi: 10.1371/journal.pone.0197541. eCollection 2018. PLoS One. 2018. PMID: 29768499 Free PMC article.
-
Ultrastructure expansion microscopy in Trypanosoma brucei.Open Biol. 2021 Oct;11(10):210132. doi: 10.1098/rsob.210132. Epub 2021 Oct 13. Open Biol. 2021. PMID: 34637654 Free PMC article.
-
Kinetoplast Division Factors in a Trypanosome.Trends Parasitol. 2019 Feb;35(2):119-128. doi: 10.1016/j.pt.2018.11.002. Epub 2019 Jan 10. Trends Parasitol. 2019. PMID: 30638954 Free PMC article. Review.
-
Cell cycle localization dynamics of mitochondrial DNA polymerase IC in African trypanosomes.Mol Biol Cell. 2018 Oct 15;29(21):2540-2552. doi: 10.1091/mbc.E18-02-0127. Epub 2018 Aug 22. Mol Biol Cell. 2018. PMID: 30133333 Free PMC article.
-
Characterization of the novel mitochondrial genome replication factor MiRF172 in Trypanosoma brucei.J Cell Sci. 2018 Apr 25;131(8):jcs211730. doi: 10.1242/jcs.211730. J Cell Sci. 2018. PMID: 29626111 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

