Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression
- PMID: 27168937
- PMCID: PMC4860271
- DOI: 10.1002/reg2.37
Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression
Abstract
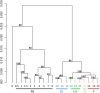
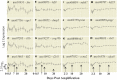
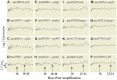
While it is appreciated that global gene expression analyses can provide novel insights about complex biological processes, experiments are generally insufficiently powered to achieve this goal. Here we report the results of a robust microarray experiment of axolotl forelimb regeneration. At each of 20 post-amputation time points, we estimated gene expression for 10 replicate RNA samples that were isolated from 1 mm of heterogeneous tissue collected from the distal limb tip. We show that the limb transcription program diverges progressively with time from the non-injured state, and divergence among time adjacent samples is mostly gradual. However, punctuated episodes of transcription were identified for five intervals of time, with four of these coinciding with well-described stages of limb regeneration-amputation, early bud, late bud, and pallet. The results suggest that regeneration is highly temporally structured and regulated by mechanisms that function within narrow windows of time to coordinate transcription within and across cell types of the regenerating limb. Our results provide an integrative framework for hypothesis generation using this complex and highly informative data set.
Keywords: Axolotl; limb; microarray; regeneration; transcription.
Figures





References
-
- Benjamini, Y. & Hochberg, Y. (1995). Journal of the Royal Statistical Society, Series B (Methodological), 57, 289–300.
-
- Carlson, B.M. (2007). Principles of Regenerative Biology. Academic; Press. Burlington, MA.
-
- Dennis, G. Jr , Sherman, B.T. , Hosack, D.A. , Yang, J. , Gao, W. , Lane, H.C. et al. (2003). DAVID: Database for annotation, visualization, and integrated discovery. Genom Biol, 4, P3. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
