RNA-Seq of Huntington's disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation
- PMID: 27170315
- PMCID: PMC5181590
- DOI: 10.1093/hmg/ddw142
RNA-Seq of Huntington's disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation
Abstract
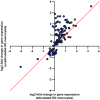
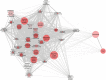
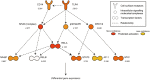
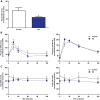
Innate immune activation beyond the central nervous system is emerging as a vital component of the pathogenesis of neurodegeneration. Huntington's disease (HD) is a fatal neurodegenerative disorder caused by a CAG repeat expansion in the huntingtin gene. The systemic innate immune system is thought to act as a modifier of disease progression; however, the molecular mechanisms remain only partially understood. Here we use RNA-sequencing to perform whole transcriptome analysis of primary monocytes from thirty manifest HD patients and thirty-three control subjects, cultured with and without a proinflammatory stimulus. In contrast with previous studies that have required stimulation to elicit phenotypic abnormalities, we demonstrate significant transcriptional differences in HD monocytes in their basal, unstimulated state. This includes previously undetected increased resting expression of genes encoding numerous proinflammatory cytokines, such as IL6 Further pathway analysis revealed widespread resting enrichment of proinflammatory functional gene sets, while upstream regulator analysis coupled with Western blotting suggests that abnormal basal activation of the NFĸB pathway plays a key role in mediating these transcriptional changes. That HD myeloid cells have a proinflammatory phenotype in the absence of stimulation is consistent with a priming effect of mutant huntingtin, whereby basal dysfunction leads to an exaggerated inflammatory response once a stimulus is encountered. These data advance our understanding of mutant huntingtin pathogenesis, establish resting myeloid cells as a key source of HD immune dysfunction, and further demonstrate the importance of systemic immunity in the potential treatment of HD and the wider study of neurodegeneration.
© The Author 2016. Published by Oxford University Press.
Figures
References
-
- The Huntington’s Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell, 72, 971–983. - PubMed
-
- Ross C.A., Tabrizi S.J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol., 10, 83–98. - PubMed
-
- Li S.H., Schilling G., Young W.S., Li X.J., Margolis R.L., Stine O.C., Wagster M.V., Abbott M.H., Franz M.L., Ranen N.G. (1993) Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron, 11, 985–993. - PubMed
-
- van der Burg J.M., Björkqvist M., Brundin P. (2009) Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol., 8, 765–774. - PubMed
-
- Tai Y.F., Pavese N., Gerhard A., Tabrizi S.J., Barker R.A., Brooks D.J., Piccini P. (2007) Microglial activation in presymptomatic Huntington's disease gene carriers. Brain, 130, 1759–1766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

