Bacterial pathogenesis of plants: future challenges from a microbial perspective: Challenges in Bacterial Molecular Plant Pathology
- PMID: 27170435
- PMCID: PMC6638335
- DOI: 10.1111/mpp.12427
Bacterial pathogenesis of plants: future challenges from a microbial perspective: Challenges in Bacterial Molecular Plant Pathology
Abstract
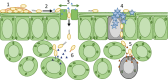
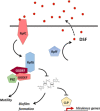
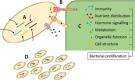
Plant infection is a complicated process. On encountering a plant, pathogenic microorganisms must first adapt to life on the epiphytic surface, and survive long enough to initiate an infection. Responsiveness to the environment is critical throughout infection, with intracellular and community-level signal transduction pathways integrating environmental signals and triggering appropriate responses in the bacterial population. Ultimately, phytopathogens must migrate from the epiphytic surface into the plant tissue using motility and chemotaxis pathways. This migration is coupled with overcoming the physical and chemical barriers to entry into the plant apoplast. Once inside the plant, bacteria use an array of secretion systems to release phytotoxins and protein effectors that fulfil diverse pathogenic functions (Fig. ) (Melotto and Kunkel, ; Phan Tran et al., ). As our understanding of the pathways and mechanisms underpinning plant pathogenicity increases, a number of central research challenges are emerging that will profoundly shape the direction of research in the future. We need to understand the bacterial phenotypes that promote epiphytic survival and surface adaptation in pathogenic bacteria. How do these pathways function in the context of the plant-associated microbiome, and what impact does this complex microbial community have on the onset and severity of plant infections? The huge importance of bacterial signal transduction to every stage of plant infection is becoming increasingly clear. However, there is a great deal to learn about how these signalling pathways function in phytopathogenic bacteria, and the contribution they make to various aspects of plant pathogenicity. We are increasingly able to explore the structural and functional diversity of small-molecule natural products from plant pathogens. We need to acquire a much better understanding of the production, deployment, functional redundancy and physiological roles of these molecules. Type III secretion systems (T3SSs) are important and well-studied contributors to bacterial disease. Several key unanswered questions will shape future investigations of these systems. We need to define the mechanism of hierarchical and temporal control of effector secretion. For successful infection, effectors need to interact with host components to exert their function. Advanced biochemical, proteomic and cell biological techniques will enable us to study the function of effectors inside the host cell in more detail and on a broader scale. Population genomics analyses provide insight into evolutionary adaptation processes of phytopathogens. The determination of the diversity and distribution of type III effectors (T3Es) and other virulence genes within and across pathogenic species, pathovars and strains will allow us to understand how pathogens adapt to specific hosts, the evolutionary pathways available to them, and the possible future directions of the evolutionary arms race between effectors and molecular plant targets. Although pathogenic bacteria employ a host of different virulence and proliferation strategies, as a result of the space constraints, this review focuses mainly on the hemibiotrophic pathogens. We discuss the process of plant infection from the perspective of these important phytopathogens, and highlight new approaches to address the outstanding challenges in this important and fast-moving field.
Keywords: Pseudomonas; Xanthomonas; bacterial signalling; phytotoxins; plant infection; plant pathogenicity; type III effectors.
© 2016 The Authors. Molecular Plant Pathology Published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures

References
-
- Allesen‐Holm, M. , Barken, K.B. , Yang, L. , Klausen, M. , Webb, J.S. , Kjelleberg, S. , Molin, S. , Givskov, M. and Tolker‐Nielsen, T. (2006) A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128. - PubMed
-
- Almeida, N.F. , Yan, S. , Lindeberg, M. , Studholme, D.J. , Schneider, D.J. , Condon, B. , Liu, H. , Viana, C.J. , Warren, A. , Evans, C. , Kemen, E. , Maclean, D. , Angot, A. , Martin, G.B. , Jones, J.D. , Collmer, A. , Setubal, J.C. and Vinatzer, B.A. (2009) A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol. Plant–Microbe Interact. 22, 52–62. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

