HIV-1 Vpu Antagonizes CD317/Tetherin by Adaptor Protein-1-Mediated Exclusion from Virus Assembly Sites
- PMID: 27170757
- PMCID: PMC4944280
- DOI: 10.1128/JVI.00504-16
HIV-1 Vpu Antagonizes CD317/Tetherin by Adaptor Protein-1-Mediated Exclusion from Virus Assembly Sites
Abstract
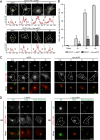
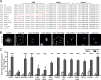
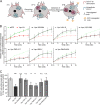
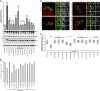
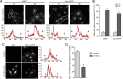
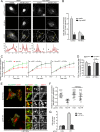
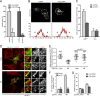
The host cell restriction factor CD317/tetherin traps virions at the surface of producer cells to prevent their release. The HIV-1 accessory protein Vpu antagonizes this restriction. Vpu reduces the cell surface density of the restriction factor and targets it for degradation; however, these activities are dispensable for enhancing particle release. Instead, Vpu has been suggested to antagonize CD317/tetherin by preventing recycling of internalized CD317/tetherin to the cell surface, blocking anterograde transport of newly synthesized CD317/tetherin, and/or displacing the restriction factor from virus assembly sites at the plasma membrane. At the molecular level, antagonism relies on the physical interaction of Vpu with CD317/tetherin. Recent findings suggested that phosphorylation of a diserine motif enables Vpu to bind to adaptor protein 1 (AP-1) trafficking complexes via two independent interaction motifs and to couple CD317/tetherin to the endocytic machinery. Here, we used a panel of Vpu proteins with specific mutations in individual interaction motifs to define which interactions are required for antagonism of CD317/tetherin. Impairing recycling or anterograde transport of CD317/tetherin to the plasma membrane was insufficient for antagonism. In contrast, excluding CD317/tetherin from HIV-1 assembly sites depended on Vpu motifs for interaction with AP-1 and CD317/tetherin and correlated with antagonism of the particle release restriction. Consistently, interference with AP-1 function or its expression blocked these Vpu activities. Our results define displacement from HIV-1 assembly sites as active principle of CD317/tetherin antagonism by Vpu and support a role of tripartite complexes between Vpu, AP-1, and CD317/tetherin in this process.
Importance: CD317/tetherin poses an intrinsic barrier to human immunodeficiency virus type 1 (HIV-1) replication in human cells by trapping virus particles at the surface of producer cells and thereby preventing their release. The viral protein Vpu antagonizes this restriction, and molecular interactions with the restriction factor and adaptor protein complex 1 (AP-1) were suggested to mediate this activity. Vpu modulates intracellular trafficking of CD317/tetherin and excludes the restriction factor from HIV-1 assembly sites at the plasma membrane, but the relative contribution of these effects to antagonism remain elusive. Using a panel of Vpu mutants, as well as interference with AP-1 function and expression, we show here that Vpu antagonizes CD317/tetherin by blocking its recruitment to viral assembly sites in an AP-1-dependent manner. These results refine our understanding of the molecular mechanisms of CD317/tetherin antagonism and suggest complexes of Vpu with the restriction factor and AP-1 as targets for potential therapeutic intervention.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
HIV-1 Vpu blocks recycling and biosynthetic transport of the intrinsic immunity factor CD317/tetherin to overcome the virion release restriction.mBio. 2011 May 24;2(3):e00036-11. doi: 10.1128/mBio.00036-11. Print 2011. mBio. 2011. PMID: 21610122 Free PMC article.
-
A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction.PLoS Pathog. 2012;8(3):e1002609. doi: 10.1371/journal.ppat.1002609. Epub 2012 Mar 29. PLoS Pathog. 2012. PMID: 22479182 Free PMC article.
-
HIV-1 Vpu's lipid raft association is dispensable for counteraction of the particle release restriction imposed by CD317/Tetherin.Virology. 2012 Mar 1;424(1):33-44. doi: 10.1016/j.virol.2011.12.008. Epub 2012 Jan 4. Virology. 2012. PMID: 22222210
-
Sites of action of HIV-1 Vpu in BST-2/tetherin downregulation.Curr HIV Res. 2012 Jun;10(4):283-91. doi: 10.2174/157016212800792423. Curr HIV Res. 2012. PMID: 22524176 Review.
-
Emerging role of the host restriction factor tetherin in viral immune sensing.J Mol Biol. 2013 Dec 13;425(24):4956-64. doi: 10.1016/j.jmb.2013.09.029. Epub 2013 Sep 26. J Mol Biol. 2013. PMID: 24075872 Review.
Cited by
-
Host Cell Restriction Factors Blocking Efficient Vector Transduction: Challenges in Lentiviral and Adeno-Associated Vector Based Gene Therapies.Cells. 2023 Feb 24;12(5):732. doi: 10.3390/cells12050732. Cells. 2023. PMID: 36899868 Free PMC article. Review.
-
Conserved residues within the HIV-1 Vpu transmembrane-proximal hinge region modulate BST2 binding and antagonism.Retrovirology. 2017 Mar 14;14(1):18. doi: 10.1186/s12977-017-0345-6. Retrovirology. 2017. PMID: 28288652 Free PMC article.
-
Substitutions in Nef That Uncouple Tetherin and SERINC5 Antagonism Impair Simian Immunodeficiency Virus Replication in Primary Rhesus Macaque Lymphocytes.J Virol. 2022 Jun 8;96(11):e0017622. doi: 10.1128/jvi.00176-22. Epub 2022 May 10. J Virol. 2022. PMID: 35536019 Free PMC article.
-
Extracellular vesicles of the blood-brain barrier: Role in the HIV-1 associated amyloid beta pathology.Mol Cell Neurosci. 2017 Mar;79:12-22. doi: 10.1016/j.mcn.2016.12.006. Epub 2016 Dec 29. Mol Cell Neurosci. 2017. PMID: 28040512 Free PMC article.
-
Tetherin Antagonism by HIV-1 Group M Nef Proteins.J Virol. 2016 Nov 14;90(23):10701-10714. doi: 10.1128/JVI.01465-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27654287 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

