Arteriviruses, Pegiviruses, and Lentiviruses Are Common among Wild African Monkeys
- PMID: 27170760
- PMCID: PMC4944300
- DOI: 10.1128/JVI.00573-16
Arteriviruses, Pegiviruses, and Lentiviruses Are Common among Wild African Monkeys
Abstract
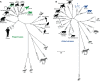
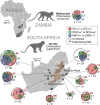
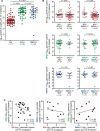
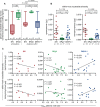
Nonhuman primates (NHPs) are a historically important source of zoonotic viruses and are a gold-standard model for research on many human pathogens. However, with the exception of simian immunodeficiency virus (SIV) (family Retroviridae), the blood-borne viruses harbored by these animals in the wild remain incompletely characterized. Here, we report the discovery and characterization of two novel simian pegiviruses (family Flaviviridae) and two novel simian arteriviruses (family Arteriviridae) in wild African green monkeys from Zambia (malbroucks [Chlorocebus cynosuros]) and South Africa (vervet monkeys [Chlorocebus pygerythrus]). We examine several aspects of infection, including viral load, genetic diversity, evolution, and geographic distribution, as well as host factors such as age, sex, and plasma cytokines. In combination with previous efforts to characterize blood-borne RNA viruses in wild primates across sub-Saharan Africa, these discoveries demonstrate that in addition to SIV, simian pegiviruses and simian arteriviruses are widespread and prevalent among many African cercopithecoid (i.e., Old World) monkeys.
Importance: Primates are an important source of viruses that infect humans and serve as an important laboratory model of human virus infection. Here, we discover two new viruses in African green monkeys from Zambia and South Africa. In combination with previous virus discovery efforts, this finding suggests that these virus types are widespread among African monkeys. Our analysis suggests that one of these virus types, the simian arteriviruses, may have the potential to jump between different primate species and cause disease. In contrast, the other virus type, the pegiviruses, are thought to reduce the disease caused by human immunodeficiency virus (HIV) in humans. However, we did not observe a similar protective effect in SIV-infected African monkeys coinfected with pegiviruses, possibly because SIV causes little to no disease in these hosts.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Grants and funding
- T32 GM081061/GM/NIGMS NIH HHS/United States
- R01 AI084787/AI/NIAID NIH HHS/United States
- T32 GM008692/GM/NIGMS NIH HHS/United States
- P51 OD011104/OD/NIH HHS/United States
- P01 AI088564/AI/NIAID NIH HHS/United States
- R01 AI116382/AI/NIAID NIH HHS/United States
- T32 AI055397/AI/NIAID NIH HHS/United States
- R01 AI045901/AI/NIAID NIH HHS/United States
- R01 TW009237/TW/FIC NIH HHS/United States
- C06 RR020141/RR/NCRR NIH HHS/United States
- C06 RR015459/RR/NCRR NIH HHS/United States
- P51 RR000167/RR/NCRR NIH HHS/United States
- R01 OD010980/OD/NIH HHS/United States
- R01 AI077376/AI/NIAID NIH HHS/United States
- HHSN272200700016I/AI/NIAID NIH HHS/United States
- R01 RR016300/RR/NCRR NIH HHS/United States
- R01 RR025781/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

