Stalled replication forks within heterochromatin require ATRX for protection
- PMID: 27171262
- PMCID: PMC4917659
- DOI: 10.1038/cddis.2016.121
Stalled replication forks within heterochromatin require ATRX for protection
Abstract
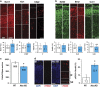
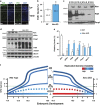
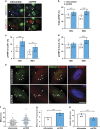
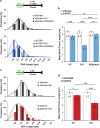
Expansive growth of neural progenitor cells (NPCs) is a prerequisite to the temporal waves of neuronal differentiation that generate the six-layered neocortex, while also placing a heavy burden on proteins that regulate chromatin packaging and genome integrity. This problem is further reflected by the growing number of developmental disorders caused by mutations in chromatin regulators. ATRX gene mutations cause a severe intellectual disability disorder (α-thalassemia mental retardation X-linked (ATRX) syndrome; OMIM no. 301040), characterized by microcephaly, urogenital abnormalities and α-thalassemia. Although the ATRX protein is required for the maintenance of repetitive DNA within heterochromatin, how this translates to disease pathogenesis remain poorly understood and was a focus of this study. We demonstrate that Atrx(FoxG1Cre) forebrain-specific conditional knockout mice display poly(ADP-ribose) polymerase-1 (Parp-1) hyperactivation during neurogenesis and generate fewer late-born Cux1- and Brn2-positive neurons that accounts for the reduced cortical size. Moreover, DNA damage, induced Parp-1 and Atm activation is elevated in progenitor cells and contributes to their increased level of cell death. ATRX-null HeLa cells are similarly sensitive to hydroxyurea-induced replication stress, accumulate DNA damage and proliferate poorly. Impaired BRCA1-RAD51 colocalization and PARP-1 hyperactivation indicated that stalled replication forks are not efficiently protected. DNA fiber assays confirmed that MRE11 degradation of stalled replication forks was rampant in the absence of ATRX or DAXX. Indeed, fork degradation in ATRX-null cells could be attenuated by treatment with the MRE11 inhibitor mirin, or exacerbated by inhibiting PARP-1 activity. Taken together, these results suggest that ATRX is required to limit replication stress during cellular proliferation, whereas upregulation of PARP-1 activity functions as a compensatory mechanism to protect stalled forks, limiting genomic damage, and facilitating late-born neuron production.
Figures


References
-
- Kleefstra T, Schenck A, Kramer JM, van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology 2014; 80: 83–94. - PubMed
-
- Gibbons RJ, Higgs DR. Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet 2000; 97: 204–212. - PubMed
-
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome). Cell 1995; 80: 837–845. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

