Generating Gene Ontology-Disease Inferences to Explore Mechanisms of Human Disease at the Comparative Toxicogenomics Database
- PMID: 27171405
- PMCID: PMC4865041
- DOI: 10.1371/journal.pone.0155530
Generating Gene Ontology-Disease Inferences to Explore Mechanisms of Human Disease at the Comparative Toxicogenomics Database
Abstract
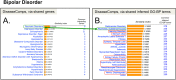
Strategies for discovering common molecular events among disparate diseases hold promise for improving understanding of disease etiology and expanding treatment options. One technique is to leverage curated datasets found in the public domain. The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) manually curates chemical-gene, chemical-disease, and gene-disease interactions from the scientific literature. The use of official gene symbols in CTD interactions enables this information to be combined with the Gene Ontology (GO) file from NCBI Gene. By integrating these GO-gene annotations with CTD's gene-disease dataset, we produce 753,000 inferences between 15,700 GO terms and 4,200 diseases, providing opportunities to explore presumptive molecular underpinnings of diseases and identify biological similarities. Through a variety of applications, we demonstrate the utility of this novel resource. As a proof-of-concept, we first analyze known repositioned drugs (e.g., raloxifene and sildenafil) and see that their target diseases have a greater degree of similarity when comparing GO terms vs. genes. Next, a computational analysis predicts seemingly non-intuitive diseases (e.g., stomach ulcers and atherosclerosis) as being similar to bipolar disorder, and these are validated in the literature as reported co-diseases. Additionally, we leverage other CTD content to develop testable hypotheses about thalidomide-gene networks to treat seemingly disparate diseases. Finally, we illustrate how CTD tools can rank a series of drugs as potential candidates for repositioning against B-cell chronic lymphocytic leukemia and predict cisplatin and the small molecule inhibitor JQ1 as lead compounds. The CTD dataset is freely available for users to navigate pathologies within the context of extensive biological processes, molecular functions, and cellular components conferred by GO. This inference set should aid researchers, bioinformaticists, and pharmaceutical drug makers in finding commonalities in disease mechanisms, which in turn could help identify new therapeutics, new indications for existing pharmaceuticals, potential disease comorbidities, and alerts for side effects.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

