A Novel Method for Gene-Specific Enhancement of Protein Translation by Targeting 5'UTRs of Selected Tumor Suppressors
- PMID: 27171412
- PMCID: PMC4865139
- DOI: 10.1371/journal.pone.0155359
A Novel Method for Gene-Specific Enhancement of Protein Translation by Targeting 5'UTRs of Selected Tumor Suppressors
Abstract
Background: Translational control is a mechanism of protein synthesis regulation emerging as an important target for new therapeutics. Naturally occurring microRNAs and synthetic small inhibitory RNAs (siRNAs) are the most recognized regulatory molecules acting via RNA interference. Surprisingly, recent studies have shown that interfering RNAs may also activate gene transcription via the newly discovered phenomenon of small RNA-induced gene activation (RNAa). Thus far, the small activating RNAs (saRNAs) have only been demonstrated as promoter-specific transcriptional activators.
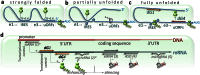
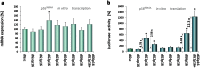
Findings: We demonstrate that oligonucleotide-based trans-acting factors can also specifically enhance gene expression at the level of protein translation by acting at sequence-specific targets within the messenger RNA 5'-untranslated region (5'UTR). We designed a set of short synthetic oligonucleotides (dGoligos), specifically targeting alternatively spliced 5'UTRs in transcripts expressed from the THRB and CDKN2A suppressor genes. The in vitro translation efficiency of reporter constructs containing alternative TRβ1 5'UTRs was increased by up to more than 55-fold following exposure to specific dGoligos. Moreover, we found that the most folded 5'UTR has higher translational regulatory potential when compared to the weakly folded TRβ1 variant. This suggests such a strategy may be especially applied to enhance translation from relatively inactive transcripts containing long 5'UTRs of complex structure.
Significance: This report represents the first method for gene-specific translation enhancement using selective trans-acting factors designed to target specific 5'UTR cis-acting elements. This simple strategy may be developed further to complement other available methods for gene expression regulation including gene silencing. The dGoligo-mediated translation-enhancing approach has the potential to be transferred to increase the translation efficiency of any suitable target gene and may have future application in gene therapy strategies to enhance expression of proteins including tumor suppressors.
Conflict of interest statement
Figures





Similar articles
-
Novel splice variants in the 5'UTR of Gtf2i expressed in the rat brain: alternative 5'UTRs and differential expression in the neuronal dendrites.J Neurochem. 2015 Aug;134(3):578-89. doi: 10.1111/jnc.13136. Epub 2015 May 14. J Neurochem. 2015. PMID: 25913238
-
Evolution of alternative and constitutive regions of mammalian 5'UTRs.BMC Genomics. 2009 Apr 16;10:162. doi: 10.1186/1471-2164-10-162. BMC Genomics. 2009. PMID: 19371439 Free PMC article.
-
Migration of Small Ribosomal Subunits on the 5' Untranslated Regions of Capped Messenger RNA.Int J Mol Sci. 2019 Sep 10;20(18):4464. doi: 10.3390/ijms20184464. Int J Mol Sci. 2019. PMID: 31510048 Free PMC article.
-
The Functional Meaning of 5'UTR in Protein-Coding Genes.Int J Mol Sci. 2023 Feb 3;24(3):2976. doi: 10.3390/ijms24032976. Int J Mol Sci. 2023. PMID: 36769304 Free PMC article. Review.
-
Targeting the Iron-Response Elements of the mRNAs for the Alzheimer's Amyloid Precursor Protein and Ferritin to Treat Acute Lead and Manganese Neurotoxicity.Int J Mol Sci. 2019 Feb 25;20(4):994. doi: 10.3390/ijms20040994. Int J Mol Sci. 2019. PMID: 30823541 Free PMC article. Review.
Cited by
-
Fatty Acids of CLA-Enriched Egg Yolks Can Induce Transcriptional Activation of Peroxisome Proliferator-Activated Receptors in MCF-7 Breast Cancer Cells.PPAR Res. 2017;2017:2865283. doi: 10.1155/2017/2865283. Epub 2017 Mar 26. PPAR Res. 2017. PMID: 28458685 Free PMC article.
-
Link between short tandem repeats and translation initiation site selection.Hum Genomics. 2018 Oct 29;12(1):47. doi: 10.1186/s40246-018-0181-3. Hum Genomics. 2018. PMID: 30373661 Free PMC article.
-
SMAD4-201 transcript as a putative biomarker in colorectal cancer.BMC Cancer. 2022 Jan 16;22(1):72. doi: 10.1186/s12885-022-09186-z. BMC Cancer. 2022. PMID: 35034624 Free PMC article.
-
Tandem repeats ubiquitously flank and contribute to translation initiation sites.BMC Genom Data. 2022 Jul 27;23(1):59. doi: 10.1186/s12863-022-01075-5. BMC Genom Data. 2022. PMID: 35896982 Free PMC article.
-
Micro-RNA 10a Is Increased in Feline T Regulatory Cells and Increases Foxp3 Protein Expression Following In Vitro Transfection.Vet Sci. 2017 Feb 21;4(1):12. doi: 10.3390/vetsci4010012. Vet Sci. 2017. PMID: 29056671 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

