Two Components of the RNA-Directed DNA Methylation Pathway Associate with MORC6 and Silence Loci Targeted by MORC6 in Arabidopsis
- PMID: 27171427
- PMCID: PMC4865133
- DOI: 10.1371/journal.pgen.1006026
Two Components of the RNA-Directed DNA Methylation Pathway Associate with MORC6 and Silence Loci Targeted by MORC6 in Arabidopsis
Abstract
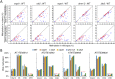
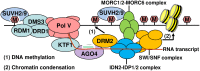
The SU(VAR)3-9 homolog SUVH9 and the double-stranded RNA-binding protein IDN2 were thought to be components of an RNA-directed DNA methylation (RdDM) pathway in Arabidopsis. We previously found that SUVH9 interacts with MORC6 but how the interaction contributes to transcriptional silencing remains elusive. Here, our genetic analysis indicates that SUVH2 and SUVH9 can either act in the same pathway as MORC6 or act synergistically with MORC6 to mediate transcriptional silencing. Moreover, we demonstrate that IDN2 interacts with MORC6 and mediates the silencing of a subset of MORC6 target loci. Like SUVH2, SUVH9, and IDN2, other RdDM components including Pol IV, Pol V, RDR2, and DRM2 are also required for transcriptional silencing at a subset of MORC6 target loci. MORC6 was previously shown to mediate transcriptional silencing through heterochromatin condensation. We demonstrate that the SWI/SNF chromatin-remodeling complex components SWI3B, SWI3C, and SWI3D interact with MORC6 as well as with SUVH9 and then mediate transcriptional silencing. These results suggest that the RdDM components are involved not only in DNA methylation but also in MORC6-mediated heterochromatin condensation. This study illustrates how DNA methylation is linked to heterochromatin condensation and thereby enhances transcriptional silencing at methylated genomic regions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




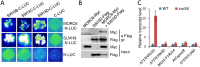
References
-
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

