HIV-1 Vpu Mediates HLA-C Downregulation
- PMID: 27173934
- PMCID: PMC4904791
- DOI: 10.1016/j.chom.2016.04.005
HIV-1 Vpu Mediates HLA-C Downregulation
Abstract
Many pathogens evade cytotoxic T lymphocytes (CTLs) by downregulating HLA molecules on infected cells, but the loss of HLA can trigger NK cell-mediated lysis. HIV-1 is thought to subvert CTLs while preserving NK cell inhibition by Nef-mediated downregulation of HLA-A and -B but not HLA-C molecules. We find that HLA-C is downregulated by most primary HIV-1 clones, including transmitted founder viruses, in contrast to the laboratory-adapted NL4-3 virus. HLA-C reduction is mediated by viral Vpu and reduces the ability of HLA-C restricted CTLs to suppress viral replication in CD4+ cells in vitro. HLA-A/B are unaffected by Vpu, and primary HIV-1 clones vary in their ability to downregulate HLA-C, possibly in response to whether CTLs or NK cells dominate immune pressure through HLA-C. HIV-2 also suppresses HLA-C expression through distinct mechanisms, underscoring the immune pressure HLA-C exerts on HIV. This viral immune evasion casts new light on the roles of CTLs and NK cells in immune responses against HIV.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

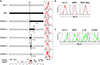

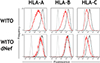

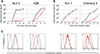

Comment in
-
HLA-C Downmodulation by HIV-1 Vpu.Cell Host Microbe. 2016 May 11;19(5):570-1. doi: 10.1016/j.chom.2016.04.023. Cell Host Microbe. 2016. PMID: 27173922
References
-
- Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leukocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127:26–39. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

