Drosophila male and female germline stem cell niches require the nuclear lamina protein Otefin
- PMID: 27174470
- PMCID: PMC4902716
- DOI: 10.1016/j.ydbio.2016.05.001
Drosophila male and female germline stem cell niches require the nuclear lamina protein Otefin
Abstract
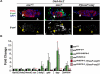
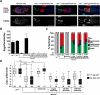
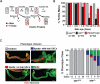
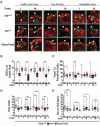
The nuclear lamina is an extensive protein network that underlies the inner nuclear envelope. This network includes the LAP2-emerin-MAN1-domain (LEM-D) protein family, proteins that share an association with the chromatin binding protein Barrier-to-autointegration factor (BAF). Loss of individual LEM-D proteins causes progressive, tissue-restricted diseases, known as laminopathies. Mechanisms associated with laminopathies are not yet understood. Here we present our studies of one of the Drosophila nuclear lamina LEM-D proteins, Otefin (Ote), a homologue of emerin. Previous studies have shown that Ote is autonomously required for the survival of female germline stem cells (GSCs). We demonstrate that Ote is also required for survival of somatic cells in the ovarian niche, with loss of Ote causing a decrease in cap cell number and altered signal transduction. We show germ cell-restricted expression of Ote rescues these defects, revealing a non-autonomous function for Ote in niche maintenance and emphasizing that GSCs contribute to the maintenance of their own niches. Further, we investigate the requirement of Ote in the male fertility. We show that ote mutant males become prematurely sterile as they age. Parallel to observations in females, this sterility is associated with GSC loss and changes in somatic cells of the niche, phenotypes that are largely rescued by germ cell-restricted Ote expression. Taken together, our studies demonstrate that Ote is required autonomously for survival of two stem cell populations, as well as non-autonomously for maintenance of two somatic niches. Finally, our data add to growing evidence that LEM-D proteins have critical roles in stem cell survival and tissue homeostasis.
Keywords: Drosophila; LEM-domain proteins; Nuclear lamina; Oogenesis, Germline stem cells; Spermatogenesis.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

