The Clathrin-dependent Spindle Proteome
- PMID: 27174698
- PMCID: PMC4974335
- DOI: 10.1074/mcp.M115.054809
The Clathrin-dependent Spindle Proteome
Abstract
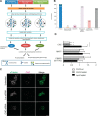
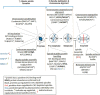
The mitotic spindle is required for chromosome congression and subsequent equal segregation of sister chromatids. These processes involve a complex network of signaling molecules located at the spindle. The endocytic protein, clathrin, has a "moonlighting" role during mitosis, whereby it stabilizes the mitotic spindle. The signaling pathways that clathrin participates in to achieve mitotic spindle stability are unknown. Here, we assessed the mitotic spindle proteome and phosphoproteome in clathrin-depleted cells using quantitative MS/MS (data are available via ProteomeXchange with identifier PXD001603). We report a spindle proteome that consists of 3046 proteins and a spindle phosphoproteome consisting of 5157 phosphosites in 1641 phosphoproteins. Of these, 2908 (95.4%) proteins and 1636 (99.7%) phosphoproteins are known or predicted spindle-associated proteins. Clathrin-depletion from spindles resulted in dysregulation of 121 proteins and perturbed signaling to 47 phosphosites. The majority of these proteins increased in mitotic spindle abundance and six of these were validated by immunofluorescence microscopy. Functional pathway analysis confirmed the reported role of clathrin in mitotic spindle stabilization for chromosome alignment and highlighted possible new mechanisms of clathrin action. The data also revealed a novel second mitotic role for clathrin in bipolar spindle formation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Musacchio A., and Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 - PubMed
-
- Sauer G., Korner R., Hanisch A., Ries A., Nigg E. A., and Sillje H. H. (2005) Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics 4, 35–43 - PubMed
-
- Compton D. A. (2000) Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114 - PubMed
-
- Sharp D. J., Rogers G. C., and Scholey J. M. (2000) Microtubule motors in mitosis. Nature 407, 41–47 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

