Learning statistical models of phenotypes using noisy labeled training data
- PMID: 27174893
- PMCID: PMC5070523
- DOI: 10.1093/jamia/ocw028
Learning statistical models of phenotypes using noisy labeled training data
Abstract
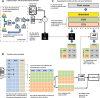
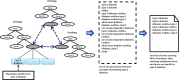
Objective: Traditionally, patient groups with a phenotype are selected through rule-based definitions whose creation and validation are time-consuming. Machine learning approaches to electronic phenotyping are limited by the paucity of labeled training datasets. We demonstrate the feasibility of utilizing semi-automatically labeled training sets to create phenotype models via machine learning, using a comprehensive representation of the patient medical record.
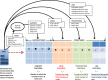
Methods: We use a list of keywords specific to the phenotype of interest to generate noisy labeled training data. We train L1 penalized logistic regression models for a chronic and an acute disease and evaluate the performance of the models against a gold standard.
Results: Our models for Type 2 diabetes mellitus and myocardial infarction achieve precision and accuracy of 0.90, 0.89, and 0.86, 0.89, respectively. Local implementations of the previously validated rule-based definitions for Type 2 diabetes mellitus and myocardial infarction achieve precision and accuracy of 0.96, 0.92 and 0.84, 0.87, respectively.We have demonstrated feasibility of learning phenotype models using imperfectly labeled data for a chronic and acute phenotype. Further research in feature engineering and in specification of the keyword list can improve the performance of the models and the scalability of the approach.
Conclusions: Our method provides an alternative to manual labeling for creating training sets for statistical models of phenotypes. Such an approach can accelerate research with large observational healthcare datasets and may also be used to create local phenotype models.
Keywords: Electronic health record; high throughput; machine learning; noisy labels; phenotyping.
© The Author 2016. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Weakly Semi-supervised phenotyping using Electronic Health records.J Biomed Inform. 2022 Oct;134:104175. doi: 10.1016/j.jbi.2022.104175. Epub 2022 Sep 5. J Biomed Inform. 2022. PMID: 36064111 Free PMC article.
-
Automated feature selection of predictors in electronic medical records data.Biometrics. 2019 Mar;75(1):268-277. doi: 10.1111/biom.12987. Epub 2019 Apr 2. Biometrics. 2019. PMID: 30353541
-
Surrogate-assisted feature extraction for high-throughput phenotyping.J Am Med Inform Assoc. 2017 Apr 1;24(e1):e143-e149. doi: 10.1093/jamia/ocw135. J Am Med Inform Assoc. 2017. PMID: 27632993 Free PMC article.
-
A review of approaches to identifying patient phenotype cohorts using electronic health records.J Am Med Inform Assoc. 2014 Mar-Apr;21(2):221-30. doi: 10.1136/amiajnl-2013-001935. Epub 2013 Nov 7. J Am Med Inform Assoc. 2014. PMID: 24201027 Free PMC article. Review.
-
Clinical Text Data in Machine Learning: Systematic Review.JMIR Med Inform. 2020 Mar 31;8(3):e17984. doi: 10.2196/17984. JMIR Med Inform. 2020. PMID: 32229465 Free PMC article. Review.
Cited by
-
Machine Learning in Rheumatic Diseases.Clin Rev Allergy Immunol. 2021 Feb;60(1):96-110. doi: 10.1007/s12016-020-08805-6. Clin Rev Allergy Immunol. 2021. PMID: 32681407 Review.
-
Predicting Future Cardiovascular Events in Patients With Peripheral Artery Disease Using Electronic Health Record Data.Circ Cardiovasc Qual Outcomes. 2019 Mar;12(3):e004741. doi: 10.1161/CIRCOUTCOMES.118.004741. Circ Cardiovasc Qual Outcomes. 2019. PMID: 30857412 Free PMC article.
-
Feature extraction for phenotyping from semantic and knowledge resources.J Biomed Inform. 2019 Mar;91:103122. doi: 10.1016/j.jbi.2019.103122. Epub 2019 Feb 7. J Biomed Inform. 2019. PMID: 30738949 Free PMC article.
-
A Roadmap for Foundational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop.Radiology. 2019 Jun;291(3):781-791. doi: 10.1148/radiol.2019190613. Epub 2019 Apr 16. Radiology. 2019. PMID: 30990384 Free PMC article.
-
Representing and utilizing clinical textual data for real world studies: An OHDSI approach.J Biomed Inform. 2023 Jun;142:104343. doi: 10.1016/j.jbi.2023.104343. Epub 2023 Mar 17. J Biomed Inform. 2023. PMID: 36935011 Free PMC article.
References
-
- Longhurst CA Harrington RA Shah NH . A ‘green button' for using aggregate patient data at the point of care .Health Aff (Millwood). 2014. ; 33 ( 7 ): 1229 – 1235 . - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

