An updated human snoRNAome
- PMID: 27174936
- PMCID: PMC4914119
- DOI: 10.1093/nar/gkw386
An updated human snoRNAome
Abstract
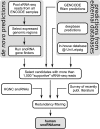
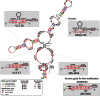
Small nucleolar RNAs (snoRNAs) are a class of non-coding RNAs that guide the post-transcriptional processing of other non-coding RNAs (mostly ribosomal RNAs), but have also been implicated in processes ranging from microRNA-dependent gene silencing to alternative splicing. In order to construct an up-to-date catalog of human snoRNAs we have combined data from various databases, de novo prediction and extensive literature review. In total, we list more than 750 curated genomic loci that give rise to snoRNA and snoRNA-like genes. Utilizing small RNA-seq data from the ENCODE project, our study characterizes the plasticity of snoRNA expression identifying both constitutively as well as cell type specific expressed snoRNAs. Especially, the comparison of malignant to non-malignant tissues and cell types shows a dramatic perturbation of the snoRNA expression profile. Finally, we developed a high-throughput variant of the reverse-transcriptase-based method for identifying 2'-O-methyl modifications in RNAs termed RimSeq. Using the data from this and other high-throughput protocols together with previously reported modification sites and state-of-the-art target prediction methods we re-estimate the snoRNA target RNA interaction network. Our current results assign a reliable modification site to 83% of the canonical snoRNAs, leaving only 76 snoRNA sequences as orphan.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

