Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape
- PMID: 27174990
- PMCID: PMC4928577
- DOI: 10.1126/science.aac7272
Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape
Abstract
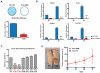
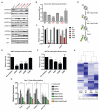
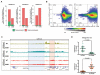
Several types of pediatric cancers reportedly contain high-frequency missense mutations in histone H3, yet the underlying oncogenic mechanism remains poorly characterized. Here we report that the H3 lysine 36-to-methionine (H3K36M) mutation impairs the differentiation of mesenchymal progenitor cells and generates undifferentiated sarcoma in vivo. H3K36M mutant nucleosomes inhibit the enzymatic activities of several H3K36 methyltransferases. Depleting H3K36 methyltransferases, or expressing an H3K36I mutant that similarly inhibits H3K36 methylation, is sufficient to phenocopy the H3K36M mutation. After the loss of H3K36 methylation, a genome-wide gain in H3K27 methylation leads to a redistribution of polycomb repressive complex 1 and de-repression of its target genes known to block mesenchymal differentiation. Our findings are mirrored in human undifferentiated sarcomas in which novel K36M/I mutations in H3.1 are identified.
Copyright © 2016, American Association for the Advancement of Science.
Figures

References
-
- Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01CA196539/CA/NCI NIH HHS/United States
- K08 CA181475/CA/NCI NIH HHS/United States
- K08CA181475/CA/NCI NIH HHS/United States
- P01 CA196539/CA/NCI NIH HHS/United States
- DP2OD007447/OD/NIH HHS/United States
- K08CA151660/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- Canadian Institutes of Health Research/Canada
- R01GM110174/GM/NIGMS NIH HHS/United States
- P30CA008748/CA/NCI NIH HHS/United States
- DP2 OD007447/OD/NIH HHS/United States
- R01 GM110174/GM/NIGMS NIH HHS/United States
- DP2CA174499/CA/NCI NIH HHS/United States
- DP2 CA174499/CA/NCI NIH HHS/United States
- R01 GM107047/GM/NIGMS NIH HHS/United States
- K08 CA151660/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

