Improved Placement of Multi-mapping Small RNAs
- PMID: 27175019
- PMCID: PMC4938663
- DOI: 10.1534/g3.116.030452
Improved Placement of Multi-mapping Small RNAs
Abstract
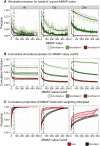
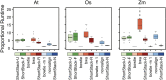
High-throughput sequencing of small RNAs (sRNA-seq) is a popular method used to discover and annotate microRNAs (miRNAs), endogenous short interfering RNAs (siRNAs), and Piwi-associated RNAs (piRNAs). One of the key steps in sRNA-seq data analysis is alignment to a reference genome. sRNA-seq libraries often have a high proportion of reads that align to multiple genomic locations, which makes determining their true origins difficult. Commonly used sRNA-seq alignment methods result in either very low precision (choosing an alignment at random), or sensitivity (ignoring multi-mapping reads). Here, we describe and test an sRNA-seq alignment strategy that uses local genomic context to guide decisions on proper placements of multi-mapped sRNA-seq reads. Tests using simulated sRNA-seq data demonstrated that this local-weighting method outperforms other alignment strategies using three different plant genomes. Experimental analyses with real sRNA-seq data also indicate superior performance of local-weighting methods for both plant miRNAs and heterochromatic siRNAs. The local-weighting methods we have developed are implemented as part of the sRNA-seq analysis program ShortStack, which is freely available under a general public license. Improved genome alignments of sRNA-seq data should increase the quality of downstream analyses and genome annotation efforts.
Keywords: alignment; annotation; bioinformatics; miRNA; sRNA-seq; siRNA.
Copyright © 2016 Johnson et al.
Figures






References
-
- Allen E., Xie Z., Gustafson A. M., Carrington J. C., 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. - PubMed
-
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., et al. , 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
