Midazolam regulated caspase pathway, endoplasmic reticulum stress, autophagy, and cell cycle to induce apoptosis in MA-10 mouse Leydig tumor cells
- PMID: 27175086
- PMCID: PMC4854247
- DOI: 10.2147/OTT.S101671
Midazolam regulated caspase pathway, endoplasmic reticulum stress, autophagy, and cell cycle to induce apoptosis in MA-10 mouse Leydig tumor cells
Abstract
Purpose: Midazolam is widely used as a sedative and anesthetic induction agent by modulating the different GABA receptors in the central nervous system. Studies have also shown that midazolam has an anticancer effect on various tumors. In a previous study, we found that midazolam could induce MA-10 mouse Leydig tumor cell apoptosis by activating caspase cascade. However, the detailed mechanism related to the upstream and downstream pathways of the caspase cascade, such as endoplasmic reticulum (ER) stress, autophagy, and p53 pathways plus cell cycle regulation in MA-10 mouse Leydig tumor cells, remains elusive.
Methods: Flow cytometry assay and Western blot analyses were exploited.
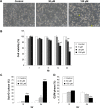
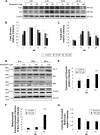
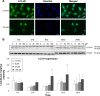
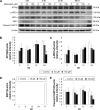
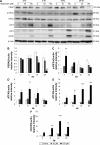
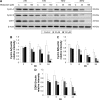
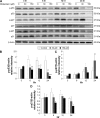
Results: Midazolam significantly decreased cell viability but increased sub-G1 phase cell numbers in MA-10 cells (P<0.05). Annexin V/propidium iodide double staining further confirmed that midazolam induced apoptosis. In addition, expressions of Fas and Fas ligand could be detected in MA-10 cells with midazolam treatments, and Bax translocation and cytochrome c release were also involved in midazolam-induced MA-10 cell apoptosis. Moreover, the staining and expression of LC3-II proteins could be observed with midazolam treatment, implying midazolam could induce autophagy to control MA-10 cell apoptosis. Furthermore, the expressions of p-EIF2α, ATF4, ATF3, and CHOP could be induced by midazolam, indicating that midazolam could stimulate apoptosis through ER stress in MA-10 cells. Additionally, the expressions of cyclin A, cyclin B, and CDK1 could be inhibited by midazolam, and the phosphorylation of p53, P27, and P21 could be adjusted by midazolam, suggesting that midazolam could manage cell cycle through the regulation of p53 pathway to induce apoptosis in MA-10 cells.
Conclusion: Midazolam could induce cell apoptosis through the activation of ER stress and the regulation of cell cycle through p53 pathway with the involvement of autophagy in MA-10 mouse Leydig tumor cells.
Keywords: ER stress; MA-10 cells; apoptosis; autophagy; caspase; cell cycle; midazolam; tumor.
Figures


References
-
- Reves JG, Fragen RJ, Vinik HR, Greenblatt DJ. Midazolam: pharmacology and uses. Anesthesiology. 1985;62(3):310–324. - PubMed
-
- García-Pedrajas F, Arroyo J. Midazolam in anesthesiology. Rev Med Univ Navarra. 1989;33(4):211–221. - PubMed
-
- Olkkola KT, Ahonen J. Midazolam and other benzodiazepines. Handb Exp Pharmacol. 2008;(182):335–360. - PubMed
-
- So EC, Chang YT, Hsing CH, et al. The effect of midazolam on mouse Leydig cell steroidogenesis and apoptosis. Toxicol Lett. 2010;192(2):169–178. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

