Peroxisome proliferator-activated receptor-gamma dependent pathway reduces the phosphorylation of dynamin-related protein 1 and ameliorates hippocampal injury induced by global ischemia in rats
- PMID: 27175924
- PMCID: PMC4865999
- DOI: 10.1186/s12929-016-0262-3
Peroxisome proliferator-activated receptor-gamma dependent pathway reduces the phosphorylation of dynamin-related protein 1 and ameliorates hippocampal injury induced by global ischemia in rats
Abstract
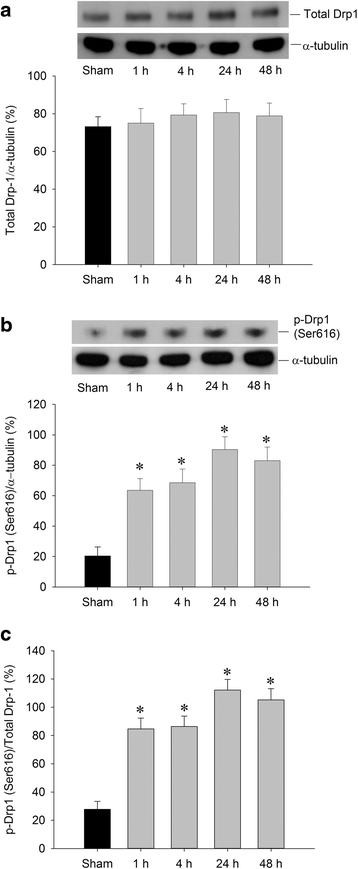
Background: Dynamin-related protein 1 (Drp1) is a mitochondrial fission protein that, upon phosphorylation at serine 616 (p-Drp1(Ser616)), plays a pivotal role in neuronal death after ischemia. In the present study, we hypothesized that peroxisome proliferator-activated receptor-gamma (PPARγ)-dependent pathway can reduce the expression of p-Drp1(Ser616) and ameliorate hippocampal injury induced by global ischemia in rats.
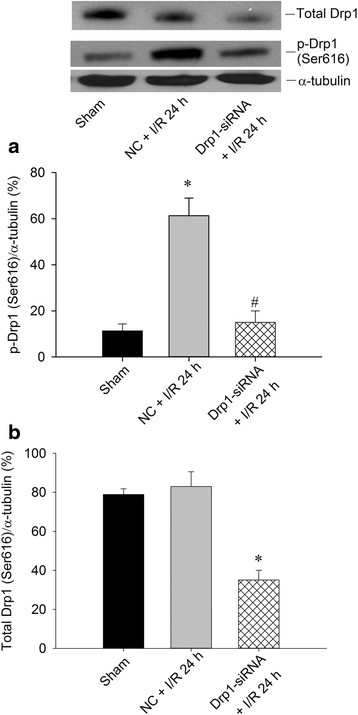
Results: We found that pretreatment of the rats with Mdivi-1, a selective Drp1 inhibitor, decreased the level of transient global ischemia (TGI)-induced p-Drp1(Ser616) and reduced cellular contents of oxidized proteins, activated caspase-3 expression as well as the extent of DNA fragmentation. Delivery of siRNA against Drp1 attenuated the expression of p-Drp1(Ser616) that was accompanied by alleviation of the TGI-induced protein oxidation, activated caspase-3 expression and DNA fragmentation in hippocampal proteins. Exogenous application of pioglitazone, a PPARγ agonist, reduced the p-Drp1(Ser616) expression, decreased TGI-induced oxidative stress and activated caspase-3 expression, lessened the extents of DNA fragmentation, and diminished the numbers of TUNEL-positive neuronal cells; all of these effects were reversed by GW9662, a PPARγ antagonist.
Conclusions: Our findings thus indicated that inhibition of TGI-induced p-Drp1(Ser616) expression by Drp1 inhibitor and Drp1-siRNA can decrease protein oxidation, activated caspase-3 expression and neuronal damage in the hippocampal CA1 subfield. PPARγ agonist, through PPARγ-dependent mechanism and via decreasing p-Drp1(Ser616) expression, can exert anti-oxidative and anti-apoptotic effects against ischemic neuronal injury.
Keywords: Apoptosis; Dynamin-related protein 1; Global ischemia; Hippocampus; Peroxisome proliferator-activated receptor-gamma; Pioglitazone.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

