Bacillus licheniformis Contains Two More PerR-Like Proteins in Addition to PerR, Fur, and Zur Orthologues
- PMID: 27176811
- PMCID: PMC4866751
- DOI: 10.1371/journal.pone.0155539
Bacillus licheniformis Contains Two More PerR-Like Proteins in Addition to PerR, Fur, and Zur Orthologues
Abstract
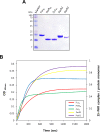
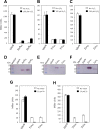
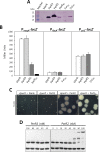
The ferric uptake regulator (Fur) family proteins include sensors of Fe (Fur), Zn (Zur), and peroxide (PerR). Among Fur family proteins, Fur and Zur are ubiquitous in most prokaryotic organisms, whereas PerR exists mainly in Gram positive bacteria as a functional homologue of OxyR. Gram positive bacteria such as Bacillus subtilis, Listeria monocytogenes and Staphylococcus aureus encode three Fur family proteins: Fur, Zur, and PerR. In this study, we identified five Fur family proteins from B. licheniformis: two novel PerR-like proteins (BL00690 and BL00950) in addition to Fur (BL05249), Zur (BL03703), and PerR (BL00075) homologues. Our data indicate that all of the five B. licheniformis Fur homologues contain a structural Zn2+ site composed of four cysteine residues like many other Fur family proteins. Furthermore, we provide evidence that the PerR-like proteins (BL00690 and BL00950) as well as PerRBL (BL00075), but not FurBL (BL05249) and ZurBL (BL03703), can sense H2O2 by histidine oxidation with different sensitivity. We also show that PerR2 (BL00690) has a PerR-like repressor activity for PerR-regulated genes in vivo. Taken together, our results suggest that B. licheniformis contains three PerR subfamily proteins which can sense H2O2 by histidine oxidation not by cysteine oxidation, in addition to Fur and Zur.
Conflict of interest statement
Figures


Similar articles
-
The difference in in vivo sensitivity between Bacillus licheniformis PerR and Bacillus subtilis PerR is due to the different cellular environments.Biochem Biophys Res Commun. 2017 Feb 26;484(1):125-131. doi: 10.1016/j.bbrc.2017.01.060. Epub 2017 Jan 16. Biochem Biophys Res Commun. 2017. PMID: 28104400
-
The inability of Bacillus licheniformis perR mutant to grow is mainly due to the lack of PerR-mediated fur repression.J Microbiol. 2017 Jun;55(6):457-463. doi: 10.1007/s12275-017-7051-x. Epub 2017 Apr 22. J Microbiol. 2017. PMID: 28434086
-
Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation.J Biol Chem. 2015 Aug 14;290(33):20374-86. doi: 10.1074/jbc.M115.664961. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134568 Free PMC article.
-
Redox Sensing by Fe2+ in Bacterial Fur Family Metalloregulators.Antioxid Redox Signal. 2018 Dec 20;29(18):1858-1871. doi: 10.1089/ars.2017.7359. Epub 2017 Oct 31. Antioxid Redox Signal. 2018. PMID: 28938859 Free PMC article. Review.
-
The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators.Arch Biochem Biophys. 2014 Mar 15;546:41-52. doi: 10.1016/j.abb.2014.01.029. Epub 2014 Feb 7. Arch Biochem Biophys. 2014. PMID: 24513162 Review.
Cited by
-
Unraveling radiation resistance strategies in two bacterial strains from the high background radiation area of Chavara-Neendakara: A comprehensive whole genome analysis.PLoS One. 2024 Jun 10;19(6):e0304810. doi: 10.1371/journal.pone.0304810. eCollection 2024. PLoS One. 2024. PMID: 38857267 Free PMC article.
-
How Microbes Defend Themselves From Incoming Hydrogen Peroxide.Front Immunol. 2021 Apr 27;12:667343. doi: 10.3389/fimmu.2021.667343. eCollection 2021. Front Immunol. 2021. PMID: 33995399 Free PMC article. Review.
-
The FUR-like regulators PerRA and PerRB integrate a complex regulatory network that promotes mammalian host-adaptation and virulence of Leptospira interrogans.PLoS Pathog. 2021 Dec 2;17(12):e1009078. doi: 10.1371/journal.ppat.1009078. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34855918 Free PMC article.
-
Comparative Genomics of Bacillus amyloliquefaciens Strains Reveals a Core Genome with Traits for Habitat Adaptation and a Secondary Metabolites Rich Accessory Genome.Front Microbiol. 2017 Aug 3;8:1438. doi: 10.3389/fmicb.2017.01438. eCollection 2017. Front Microbiol. 2017. PMID: 28824571 Free PMC article.
-
The oxidative stress response of pathogenic Leptospira is controlled by two peroxide stress regulators which putatively cooperate in controlling virulence.PLoS Pathog. 2021 Dec 2;17(12):e1009087. doi: 10.1371/journal.ppat.1009087. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34855911 Free PMC article.
References
-
- Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):5045–50. Epub 2011/03/09. 10.1073/pnas.1017744108 ; PubMed Central PMCID: PMCPmc3064357. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

