The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome
- PMID: 27177192
- PMCID: PMC5148197
- DOI: 10.1038/ismej.2016.76
The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome
Abstract
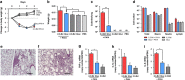
Diseases that favor colonization of the respiratory tract with Pseudomonas aeruginosa are characterized by an altered airway microbiome. Virulence of P. aeruginosa respiratory tract infection is likely influenced by interactions with other lung microbiota or their products. The bacterial fermentation product 2,3-butanediol enhances virulence and biofilm formation of P. aeruginosa in vitro. This study assessed the effects of 2,3-butanediol on P. aeruginosa persistence, inflammatory response, and the lung microbiome in vivo. Here, P. aeruginosa grown in the presence of 2,3-butanediol and encapsulated in agar beads persisted longer in the murine respiratory tract, induced enhanced TNF-α and IL-6 responses and resulted in increased colonization in the lung tissue by environmental microbes. These results led to the following hypothesis that now needs to be tested with a larger study: fermentation products from the lung microbiota not only have a role in P. aeruginosa virulence and abundance, but also on the increased colonization of the respiratory tract with environmental microbes, resulting in dynamic shifts in microbiota diversity and disease susceptibility.
Figures

Similar articles
-
Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa.ISME J. 2014 Jun;8(6):1210-20. doi: 10.1038/ismej.2013.232. Epub 2014 Jan 9. ISME J. 2014. PMID: 24401856 Free PMC article.
-
Inhibition of Pseudomonas aeruginosa secreted virulence factors reduces lung inflammation in CF mice.Virulence. 2018;9(1):1008-1018. doi: 10.1080/21505594.2018.1489198. Virulence. 2018. PMID: 29938577 Free PMC article.
-
Innate lung defenses and compromised Pseudomonas aeruginosa clearance in the malnourished mouse model of respiratory infections in cystic fibrosis.Infect Immun. 2000 Apr;68(4):2142-7. doi: 10.1128/IAI.68.4.2142-2147.2000. Infect Immun. 2000. PMID: 10722612 Free PMC article.
-
Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients.Curr Opin Pediatr. 2007 Feb;19(1):83-8. doi: 10.1097/MOP.0b013e3280123a5d. Curr Opin Pediatr. 2007. PMID: 17224667 Review.
-
The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung.FEMS Microbiol Lett. 2017 Aug 15;364(15):fnx128. doi: 10.1093/femsle/fnx128. FEMS Microbiol Lett. 2017. PMID: 28859314 Free PMC article. Review.
Cited by
-
The impact of high-IgE levels on metabolome and microbiome in experimental allergic enteritis.Allergy. 2024 Dec;79(12):3430-3447. doi: 10.1111/all.16202. Epub 2024 Jun 23. Allergy. 2024. PMID: 38932655 Free PMC article.
-
Exposure to particulate matter 2.5 leading to lung microbiome disorder and the alleviation effect of Auricularia auricular-judae polysaccharide.Int J Occup Med Environ Health. 2022 Dec 15;35(6):651-664. doi: 10.13075/ijomeh.1896.01742. Epub 2022 Jul 26. Int J Occup Med Environ Health. 2022. PMID: 35913271 Free PMC article.
-
Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes.PLoS One. 2020 Apr 21;15(4):e0231865. doi: 10.1371/journal.pone.0231865. eCollection 2020. PLoS One. 2020. PMID: 32315360 Free PMC article.
-
Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits.Genes (Basel). 2021 May 17;12(5):752. doi: 10.3390/genes12050752. Genes (Basel). 2021. PMID: 34067621 Free PMC article.
-
Differential responses of human dendritic cells to metabolites from the oral/airway microbiome.Clin Exp Immunol. 2017 Jun;188(3):371-379. doi: 10.1111/cei.12943. Epub 2017 Mar 21. Clin Exp Immunol. 2017. PMID: 28194750 Free PMC article.
References
-
- Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ et al. (2014). Antibiotic management of lung infections in cystic fibrosis. I. the microbiome, methicillin-resistant staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann Am Thorac Soc 11: 1120–1129. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

