A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs
- PMID: 27177310
- PMCID: PMC4865845
- DOI: 10.1038/ncomms11320
A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs
Abstract
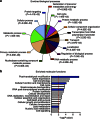
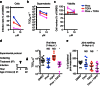
Chikungunya virus (CHIKV) is a globally spreading alphavirus against which there is no commercially available vaccine or therapy. Here we use a genome-wide siRNA screen to identify 156 proviral and 41 antiviral host factors affecting CHIKV replication. We analyse the cellular pathways in which human proviral genes are involved and identify druggable targets. Twenty-one small-molecule inhibitors, some of which are FDA approved, targeting six proviral factors or pathways, have high antiviral activity in vitro, with low toxicity. Three identified inhibitors have prophylactic antiviral effects in mouse models of chikungunya infection. Two of them, the calmodulin inhibitor pimozide and the fatty acid synthesis inhibitor TOFA, have a therapeutic effect in vivo when combined. These results demonstrate the value of loss-of-function screening and pathway analysis for the rational identification of small molecules with therapeutic potential and pave the way for the development of new, host-directed, antiviral agents.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

