Long non-coding RNAs in normal and malignant hematopoiesis
- PMID: 27177333
- PMCID: PMC5226612
- DOI: 10.18632/oncotarget.9308
Long non-coding RNAs in normal and malignant hematopoiesis
Abstract
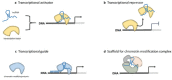
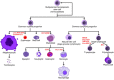
Long non-coding RNAs (lncRNAs) are defined as ncRNAs of more than 200 nt in length. They are involved in a large spectrum of biological processes, such as maintenance of genome integrity, genomic imprinting, cell differentiation, and development by means of mechanisms that remain to be fully elucidated. Besides their role in normal cellular physiology, accumulating evidence has linked lncRNA expression and functions to cancer development and progression. In this review, we summarize and discuss what is known about their expression and roles in hematopoiesis with a particular focus on their cell-type speciï¬city, functional interactions, and involvement in the pathobiology of hematological malignancies.
Keywords: hematological malignancies; hematopoiesis; long non-coding RNAs; transcriptional regulation; translation regulation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. - PMC - PubMed
-
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology. 2012;19:586–593. - PubMed
-
- Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. The Lancet Oncology. 2012;13:e249–258. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

