Domestication-driven Gossypium profilin 1 (GhPRF1) gene transduces early flowering phenotype in tobacco by spatial alteration of apical/floral-meristem related gene expression
- PMID: 27177585
- PMCID: PMC4866011
- DOI: 10.1186/s12870-016-0798-0
Domestication-driven Gossypium profilin 1 (GhPRF1) gene transduces early flowering phenotype in tobacco by spatial alteration of apical/floral-meristem related gene expression
Abstract
Background: Plant profilin genes encode core cell-wall structural proteins and are evidenced for their up-regulation under cotton domestication. Notwithstanding striking discoveries in the genetics of cell-wall organization in plants, little is explicit about the manner in which profilin-mediated molecular interplay and corresponding networks are altered, especially during cellular signalling of apical meristem determinacy and flower development.
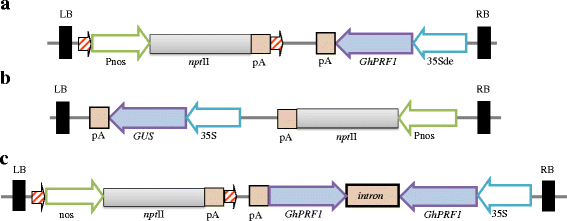
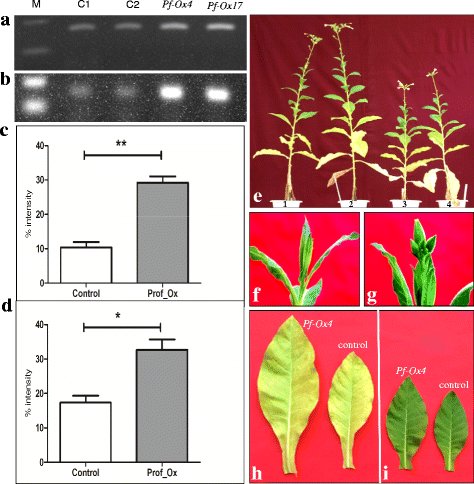
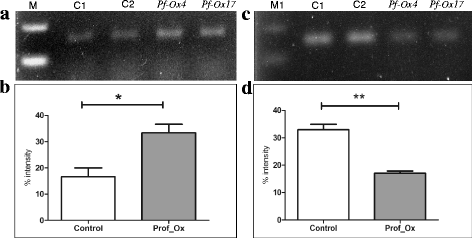
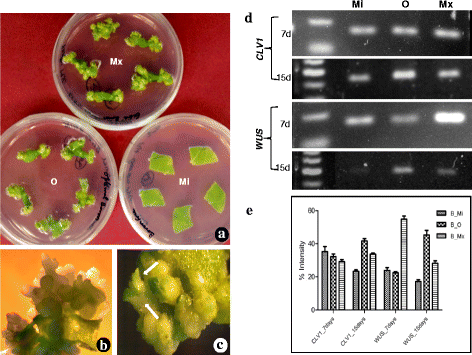
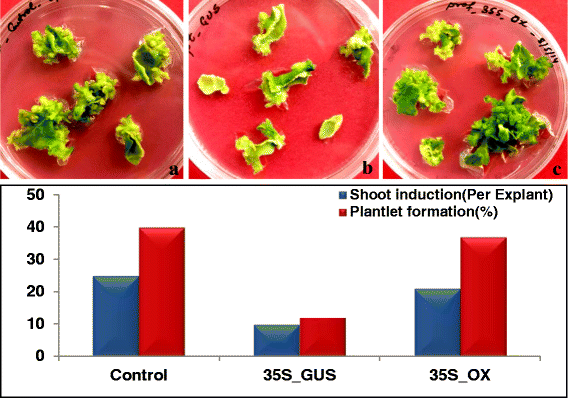
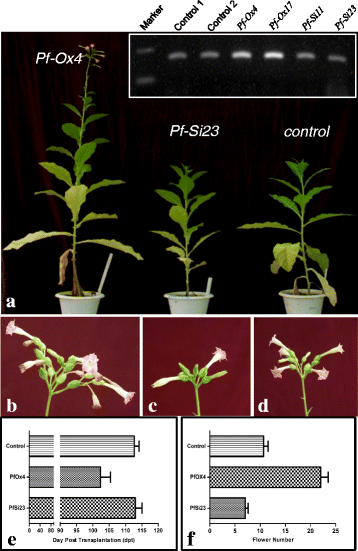
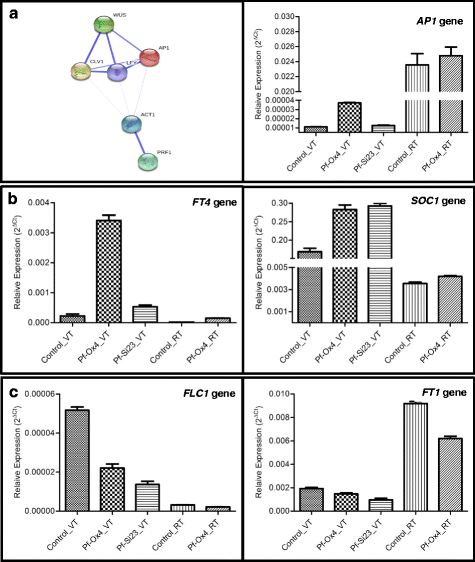
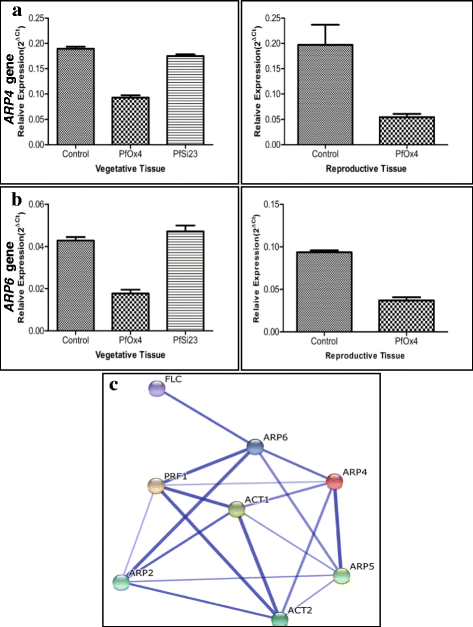
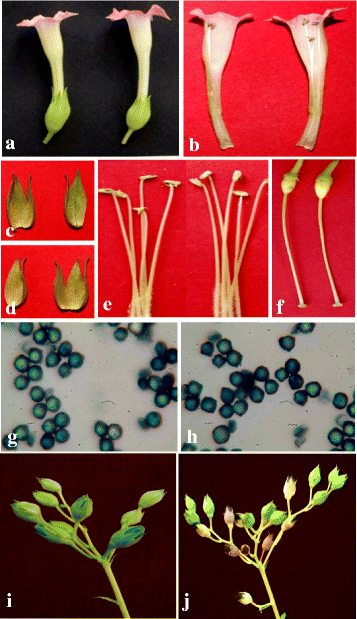
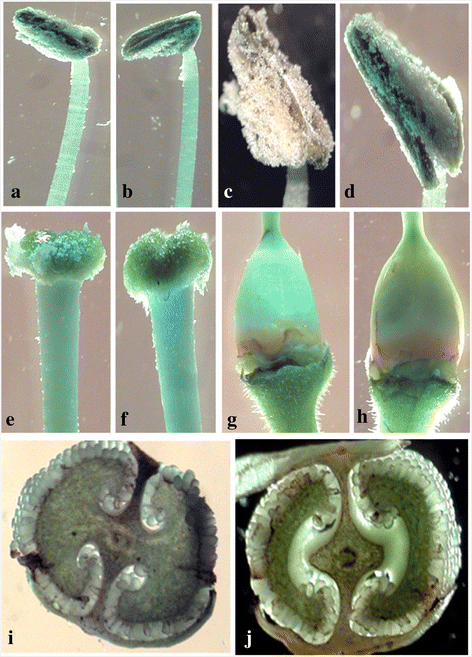
Results: Here we show that the ectopic expression of GhPRF1 gene in tobacco resulted in the hyperactivation of apical meristem and early flowering phenotype with increased flower number in comparison to the control plants. Spatial expression alteration in CLV1, a key meristem-determinacy gene, is induced by the GhPRF1 overexpression in a WUS-dependent manner and mediates cell signalling to promote flowering. But no such expression alterations are recorded in the GhPRF1-RNAi lines. The GhPRF1 transduces key positive flowering regulator AP1 gene via coordinated expression of FT4, SOC1, FLC1 and FT1 genes involved in the apical-to-floral meristem signalling cascade which is consistent with our in silico profilin interaction data. Remarkably, these positive and negative flowering regulators are spatially controlled by the Actin-Related Protein (ARP) genes, specifically ARP4 and ARP6 in proximate association with profilins. This study provides a novel and systematic link between GhPRF1 gene expression and the flower primordium initiation via up-regulation of the ARP genes, and an insight into the functional characterization of GhPRF1 gene acting upstream to the flowering mechanism. Also, the transgenic plants expressing GhPRF1 gene show an increase in the plant height, internode length, leaf size and plant vigor.
Conclusions: Overexpression of GhPRF1 gene induced early and increased flowering in tobacco with enhanced plant vigor. During apical meristem determinacy and flower development, the GhPRF1 gene directly influences key flowering regulators through ARP-genes, indicating for its role upstream in the apical-to-floral meristem signalling cascade.
Keywords: Apical meristem determinacy; Flower development; Flowering genes; Gene expression; Profilin.
Figures

Similar articles
-
Transcriptional loss of domestication-driven cytoskeletal GhPRF1 gene causes defective floral and fiber development in cotton (Gossypium).Plant Mol Biol. 2021 Dec;107(6):519-532. doi: 10.1007/s11103-021-01200-5. Epub 2021 Oct 4. Plant Mol Biol. 2021. PMID: 34606035
-
Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Co-operatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice.Plant Cell Physiol. 2017 May 1;58(5):893-903. doi: 10.1093/pcp/pcx038. Plant Cell Physiol. 2017. PMID: 28371923
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
-
AfLFY, a LEAFY homolog in Argyranthemum frutescens, controls flowering time and leaf development.Sci Rep. 2020 Jan 31;10(1):1616. doi: 10.1038/s41598-020-58570-x. Sci Rep. 2020. PMID: 32005948 Free PMC article.
-
Mechanisms and function of flower and inflorescence reversion.J Exp Bot. 2005 Oct;56(420):2587-99. doi: 10.1093/jxb/eri254. Epub 2005 Aug 30. J Exp Bot. 2005. PMID: 16131510 Review.
Cited by
-
Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation?Plants (Basel). 2022 Sep 8;11(18):2348. doi: 10.3390/plants11182348. Plants (Basel). 2022. PMID: 36145748 Free PMC article.
-
Concomitant Expression Evolution of Cell Wall Cytoskeletal Geneic Triad(s) Controls Floral Organ Shape and Fiber Emergence in Cotton (Gossypium).Front Plant Sci. 2022 May 20;13:900521. doi: 10.3389/fpls.2022.900521. eCollection 2022. Front Plant Sci. 2022. PMID: 35668801 Free PMC article. No abstract available.
-
Comparative proteomic analysis of eggplant (Solanum melongena L.) heterostylous pistil development.PLoS One. 2017 Jun 6;12(6):e0179018. doi: 10.1371/journal.pone.0179018. eCollection 2017. PLoS One. 2017. PMID: 28586360 Free PMC article.
-
Evolution of Functional Diversity Among Actin-Binding Profilin Genes in Land Plants.Front Cell Dev Biol. 2020 Dec 16;8:588689. doi: 10.3389/fcell.2020.588689. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33392185 Free PMC article. No abstract available.
-
Genome-Wide Analysis of VILLIN Gene Family Associated with Stress Responses in Cotton (Gossypium spp.).Curr Issues Mol Biol. 2024 Mar 11;46(3):2278-2300. doi: 10.3390/cimb46030146. Curr Issues Mol Biol. 2024. PMID: 38534762 Free PMC article.
References
-
- Wan JM, Jiang L, Tang JY, Wang CM, Hou MY, Jing W, Zhang LX. Genetic dissection of the seed dormancy trait in cultivated rice (Oryza sativa L.) Plant Sci. 2005;170:786–92. doi: 10.1016/j.plantsci.2005.11.011. - DOI
-
- Blumler M. Modelling the origins of legume domestication and cultivation. Economic Bot. 1991;45:243–50. doi: 10.1007/BF02862051. - DOI
-
- Plitman U, Kislev M. Reproductive changes induced by domestication. In: Stirton C, Zarucchi J, editors. Advances in legume biology. St. Louis: Missouri Botanical Garden; 1989. pp. 487–503.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

