Transcriptome analysis of dormant tomonts of the marine fish ectoparasitic ciliate Cryptocaryon irritans under low temperature
- PMID: 27177617
- PMCID: PMC4867990
- DOI: 10.1186/s13071-016-1550-1
Transcriptome analysis of dormant tomonts of the marine fish ectoparasitic ciliate Cryptocaryon irritans under low temperature
Abstract
Background: Cryptocaryon irritans, a species of obligatory ciliate ectoparasite, can infect various species of marine teleost fish. Cryptocaryon irritans that fall to the seabed or aquarium bottom in winter can form "dormant tomonts" and wake up when the temperature rises the next year. Abundant studies and analyses on the dormant tomonts were carried out at the transcriptome level, in order to investigate the molecular mechanism of C. irritans tomonts entering the dormant state under low-temperature conditions.
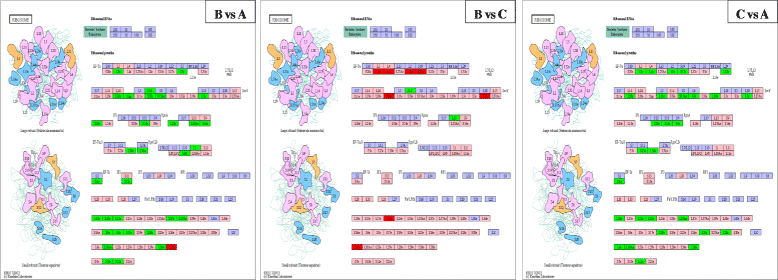
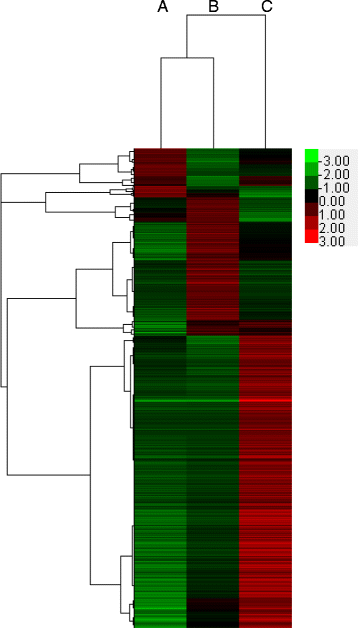
Methods: The paired-end sequencing strategy was used to better assemble the entire transcriptome de novo. All clean sequencing reads from each of the three libraries (Group A: untreated blank control; Group B: treated for 24 h at 12 °C; and Group C: developed for 24 h at 25 °C) were respectively mapped back to the transcriptome assembly using the bioinformatics software.
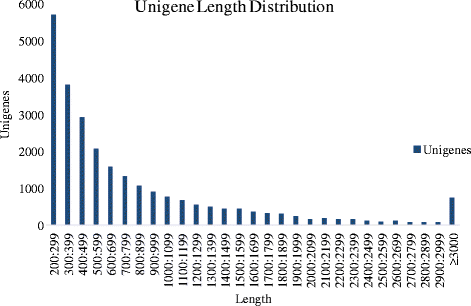
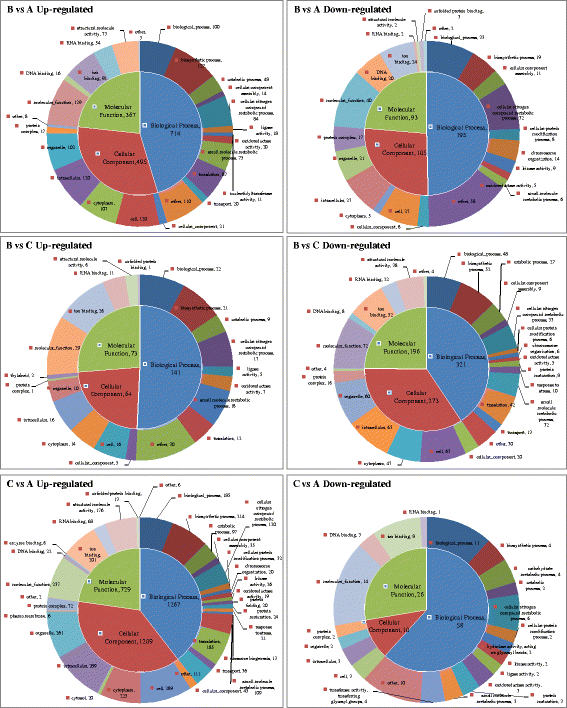
Results: In this study, 25,695,034, 21,944,467, and 28,722,875 paired-end clean reads were obtained respectively from the three cDNA libraries of the C. irritans tomont by Illumina paired-end sequencing technology. A total of 25,925 unique transcript fragments (unigenes) were assembled, with an average length of 839 bp. Differentially expressed genes (DEGs) were scrutinized; in Group B/A pairwise comparison, 343 genes presented differential expression, including 265 up-regulated genes and 78 down-regulated genes in Group B; in Group C/A pairwise comparison, there were 567 DEGs, including 548 up-regulated genes and 19 down-regulated genes in Group C; and in Group B/C pairwise comparison, 185 genes showed differential expression, including 145 up-regulated genes and 40 down-regulated genes in Group B.
Conclusions: This is the first transcriptomic analytical study of the C. irritans tomonts under low temperature. It can be concluded that most of the genes required for its cell survival under low temperature, or for cell entry into a deeper dormancy state were discovered, and that they might be considered as candidate genes to develop the diagnostic and control measures for cryptocaryoniasis.
Keywords: Cell division; Cryptocaryon irritans; Dormant tomont; Low temperature; Transcriptome.
Figures



References
-
- How KH, Zenke K, Yoshinaga T. Dynamics and distribution properties of theronts of the parasitic ciliate Cryptocaryon irritans. Aquaculture. 2015;438:170–5. doi: 10.1016/j.aquaculture.2014.12.013. - DOI
-
- Cheung PJ, Nigrelli RF, Ruggieri GD. Studies on cryptocaryoniasis in marine fish: effect of temperature and salinity on the reproductive cycle of Cryptocaryon irritans Brown, 1951. J Fish Dis. 1979;2(2):93–7. doi: 10.1111/j.1365-2761.1979.tb00146.x. - DOI
-
- Chen AP, Jiang YL, Qian D, Chen CF, Li AX, Huang J, Yang B. Cryptocaryoniasis. China Fish. 2011;8:39–40.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

