A temperature-dependent in silico model of the human ether-à-go-go-related (hERG) gene channel
- PMID: 27178106
- PMCID: PMC5042861
- DOI: 10.1016/j.vascn.2016.05.005
A temperature-dependent in silico model of the human ether-à-go-go-related (hERG) gene channel
Abstract
Introduction: Current regulatory guidelines for assessing the risk of QT prolongation include in vitro assays assessing drug effects on the human ether-à-go-go-related (hERG; also known as Kv11.1) channel expressed in cell lines. These assays are typically conducted at room temperature to promote the ease and stability of recording hERG currents. However, the new Comprehensive in vitro Proarrhythmia Assay (CiPA) paradigm proposes to use an in silico model of the human ventricular myocyte to assess risk, requiring as input hERG channel pharmacology data obtained at physiological temperatures. To accommodate current industry safety pharmacology practices for measuring hERG channel activity, an in silico model of hERG channel that allows for the extrapolation of hERG assay data across different temperatures is desired. Because temperature may have an effect on both channel gating and drug binding rate, such models may need to have two components: a base model dealing with temperature-dependent gating changes without drug, and a pharmacodynamic component simulating temperature-dependent drug binding kinetics. As a first step, a base mode that can capture temperature effects on hERG channel gating without drug is needed.
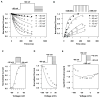
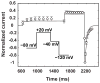
Methods and results: To meet this need for a temperature-dependent base model, a Markov model of the hERG channel with state transition rates explicitly dependent on temperature was developed and calibrated using data from a variety of published experiments conducted over a range of temperatures. The model was able to reproduce observed temperature-dependent changes in key channel gating properties and also to predict the results obtained in independent sets of new experiments.
Discussion: This new temperature-sensitive model of hERG gating represents an attempt to improve the predictivity of safety pharmacology testing by enabling the translation of room temperature hERG assay data to more physiological conditions. With further development, this model can be incorporated into the CiPA paradigm and also be used as a tool for developing insights into the thermodynamics of hERG channel gating mechanisms and the temperature-dependence of hERG channel block by drugs.
Keywords: CiPA; Kv11.1; Markov model; Methods; Temperature; hERG.
Published by Elsevier Inc.
Conflict of interest statement
Statement The authors declared no conflict of interest.
Figures



Similar articles
-
Improving the In Silico Assessment of Proarrhythmia Risk by Combining hERG (Human Ether-à-go-go-Related Gene) Channel-Drug Binding Kinetics and Multichannel Pharmacology.Circ Arrhythm Electrophysiol. 2017 Feb;10(2):e004628. doi: 10.1161/CIRCEP.116.004628. Circ Arrhythm Electrophysiol. 2017. PMID: 28202629
-
The cardiac hERG/IKr potassium channel as pharmacological target: structure, function, regulation, and clinical applications.Curr Pharm Des. 2006;12(18):2271-83. doi: 10.2174/138161206777585102. Curr Pharm Des. 2006. PMID: 16787254 Review.
-
In silico prediction of the chemical block of human ether-a-go-go-related gene (hERG) K+ current.J Physiol Sci. 2008 Dec;58(7):459-70. doi: 10.2170/physiolsci.RV-0114-08-07-R1. Epub 2008 Nov 27. J Physiol Sci. 2008. PMID: 19032804
-
An evaluation of 30 clinical drugs against the comprehensive in vitro proarrhythmia assay (CiPA) proposed ion channel panel.J Pharmacol Toxicol Methods. 2016 Sep-Oct;81:251-62. doi: 10.1016/j.vascn.2016.03.009. Epub 2016 Apr 6. J Pharmacol Toxicol Methods. 2016. PMID: 27060526
-
hERG K(+) channels: structure, function, and clinical significance.Physiol Rev. 2012 Jul;92(3):1393-478. doi: 10.1152/physrev.00036.2011. Physiol Rev. 2012. PMID: 22988594 Review.
Cited by
-
Inhibitory effects and mechanism of dihydroberberine on hERG channels expressed in HEK293 cells.PLoS One. 2017 Aug 1;12(8):e0181823. doi: 10.1371/journal.pone.0181823. eCollection 2017. PLoS One. 2017. PMID: 28763460 Free PMC article.
-
Rapid Characterization of hERG Channel Kinetics II: Temperature Dependence.Biophys J. 2019 Dec 17;117(12):2455-2470. doi: 10.1016/j.bpj.2019.07.030. Epub 2019 Jul 25. Biophys J. 2019. PMID: 31451180 Free PMC article.
-
Calibration of ionic and cellular cardiac electrophysiology models.Wiley Interdiscip Rev Syst Biol Med. 2020 Jul;12(4):e1482. doi: 10.1002/wsbm.1482. Epub 2020 Feb 21. Wiley Interdiscip Rev Syst Biol Med. 2020. PMID: 32084308 Free PMC article. Review.
-
General Principles for the Validation of Proarrhythmia Risk Prediction Models: An Extension of the CiPA In Silico Strategy.Clin Pharmacol Ther. 2020 Jan;107(1):102-111. doi: 10.1002/cpt.1647. Epub 2019 Nov 10. Clin Pharmacol Ther. 2020. PMID: 31709525 Free PMC article.
-
Electrophysiological characterization of the hERG R56Q LQTS variant and targeted rescue by the activator RPR260243.J Gen Physiol. 2021 Oct 4;153(10):e202112923. doi: 10.1085/jgp.202112923. Epub 2021 Aug 16. J Gen Physiol. 2021. PMID: 34398210 Free PMC article.
References
-
- Clancy CE, Rudy Y. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature. 1999;400:566–569. - PubMed
-
- Clancy CE, Rudy Y. Cellular consequences of HERG mutations in the long QT syndrome: precursors to sudden cardiac death. Cardiovascular Research. 2001;50:301–313. - PubMed
-
- Di Veroli GY, Davies MR, Zhang H, Abi-Gerges N, Boyett MR. High-throughput screening of drug-binding dynamics to HERG improves early drug safety assessment. American Journal of Physiology - Heart and Circulatory Physiology. 2013;304:H104–H117. - PubMed
-
- Di Veroli GY, Davies MR, Zhang H, Abi-Gerges N, Boyett MR. hERG inhibitors with similar potency but different binding kinetics do not pose the same proarrhythmic risk: implications for drug safety assessment. Journal of Cardiovascular Electrophysiology. 2014;25:197–207. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

