Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes
- PMID: 27178736
- PMCID: PMC11108369
- DOI: 10.1007/s00018-016-2271-5
Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes
Abstract
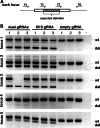
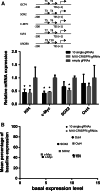
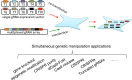
The engineered CRISPR/Cas9 technology has developed as the most efficient and broadly used genome editing tool. However, simultaneously targeting multiple genes (or genomic loci) in the same individual cells using CRISPR/Cas9 remain one technical challenge. In this article, we have developed a Golden Gate Assembly method for the generation of CRISPR gRNA expression arrays, thus enabling simultaneous gene targeting. Using this method, the generation of CRISPR gRNA expression array can be accomplished in 2 weeks, and contains up to 30 gRNA expression cassettes. We demonstrated in the study that simultaneously targeting 10 genomic loci or simultaneously inhibition of multiple endogenous genes could be achieved using the multiplexed gRNA expression array vector in human cells. The complete set of plasmids is available through the non-profit plasmid repository Addgene.
Keywords: CRISPR; Cas9; Genome editing; Golden Gate Assembly; Simultaneous multiple gene inhibition; Simultaneous multiple gene knockout.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

