Role of hippocampal activity-induced transcription in memory consolidation
- PMID: 27180338
- PMCID: PMC6526723
- DOI: 10.1515/revneuro-2016-0010
Role of hippocampal activity-induced transcription in memory consolidation
Abstract
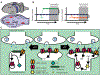
Experience-dependent changes in the strength of connections between neurons in the hippocampus (HPC) are critical for normal learning and memory consolidation, and disruption of this process drives a variety of neurological and psychiatric diseases. Proper HPC function relies upon discrete changes in gene expression driven by transcription factors (TFs) induced by neuronal activity. Here, we describe the induction and function of many of the most well-studied HPC TFs, including cyclic-AMP response element binding protein, serum-response factor, AP-1, and others, and describe their role in the learning process. We also discuss the known target genes of many of these TFs and the purported mechanisms by which they regulate long-term changes in HPC synaptic strength. Moreover, we propose that future research in this field will depend upon unbiased identification of additional gene targets for these activity-dependent TFs and subsequent meta-analyses that identify common genes or pathways regulated by multiple TFs in the HPC during learning or disease.
Figures

References
-
- Albensi BC and Mattson MP (2000). Evidence for the involvement of TNF and NF-kB in hippocampal synaptic plasticity. Synapse 35, 151–159. - PubMed
-
- Alberini CM, Ghirardl M, Metz R, and Kandel ER (1994). C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in aplysia. Cell 76, 1099–1114. - PubMed
-
- Andersson M, Westin JE, and Cenci MA (2003). Time course of striatal ΔFosB-like immunoreactivity and prodynorphin mRNA levels after discontinuation of chronic dopaminomimetic treatment. Eur. J. Neurosci 17, 661–666. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
