microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection
- PMID: 27180809
- PMCID: PMC4867432
- DOI: 10.1038/srep25997
microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection
Abstract
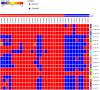
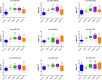
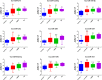
Breast cancer (BC) is the most common cancer type and the second cause of cancer-related death among women. Therefore, better understanding of breast cancer tumor biology and the identification of novel biomarkers is essential for the early diagnosis and for better disease stratification and management choices. Herein we developed a novel approach which relies on the isolation of circulating microRNAs through an enrichment step using speed-vacuum concentration which resulted in 5-fold increase in microRNA abundance. Global miRNA microarray expression profiling performed on individual samples from 23 BC and 9 normals identified 18 up-regulated miRNAs in BC patients (p(corr) < 0.05). Nine miRNAs (hsa-miR-4270, hsa-miR-1225-5p, hsa-miR-188-5p, hsa-miR-1202, hsa-miR-4281, hsa-miR-1207-5p, hsa-miR-642b-3p, hsa-miR-1290, and hsa-miR-3141) were subsequently validated using qRT-PCR in a cohort of 46 BC and 14 controls. The expression of those microRNAs was overall higher in patients with stage I, II, and III, compared to stage IV, with potential utilization for early detection. The expression of this microRNA panel was slightly higher in the HER2 and TN compared to patients with luminal subtype. Therefore, we developed a novel approach which led to the identification of a novel microRNA panel which was upregulated in BC patients with potential utilization in disease diagnosis and stratification.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

