SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b
- PMID: 27180906
- PMCID: PMC4892983
- DOI: 10.1016/j.cell.2016.04.033
SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b
Abstract
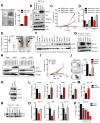
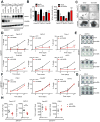
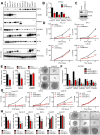
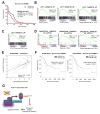
Chromatin remodeling proteins are frequently dysregulated in human cancer, yet little is known about how they control tumorigenesis. Here, we uncover an epigenetic program mediated by the NAD(+)-dependent histone deacetylase Sirtuin 6 (SIRT6) that is critical for suppression of pancreatic ductal adenocarcinoma (PDAC), one of the most lethal malignancies. SIRT6 inactivation accelerates PDAC progression and metastasis via upregulation of Lin28b, a negative regulator of the let-7 microRNA. SIRT6 loss results in histone hyperacetylation at the Lin28b promoter, Myc recruitment, and pronounced induction of Lin28b and downstream let-7 target genes, HMGA2, IGF2BP1, and IGF2BP3. This epigenetic program defines a distinct subset with a poor prognosis, representing 30%-40% of human PDAC, characterized by reduced SIRT6 expression and an exquisite dependence on Lin28b for tumor growth. Thus, we identify SIRT6 as an important PDAC tumor suppressor and uncover the Lin28b pathway as a potential therapeutic target in a molecularly defined PDAC subset. PAPERCLIP.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepato-gastroenterology. 2008;55:2016–2027. - PubMed
-
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer research. 2008;68:2587–2591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

