High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing
- PMID: 27180908
- PMCID: PMC4912470
- DOI: 10.1016/j.cell.2016.04.044
High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing
Abstract
A scalable and high-throughput method to identify precise subcellular localization of endogenous proteins is essential for integrative understanding of a cell at the molecular level. Here, we developed a simple and generalizable technique to image endogenous proteins with high specificity, resolution, and contrast in single cells in mammalian brain tissue. The technique, single-cell labeling of endogenous proteins by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-mediated homology-directed repair (SLENDR), uses in vivo genome editing to insert a sequence encoding an epitope tag or a fluorescent protein to a gene of interest by CRISPR-Cas9-mediated homology-directed repair (HDR). Single-cell, HDR-mediated genome editing was achieved by delivering the editing machinery to dividing neuronal progenitors through in utero electroporation. We demonstrate that SLENDR allows rapid determination of the localization and dynamics of many endogenous proteins in various cell types, regions, and ages in the brain. Thus, SLENDR provides a high-throughput platform to map the subcellular localization of endogenous proteins with the resolution of micro- to nanometers in the brain.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
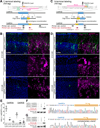
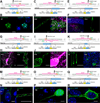
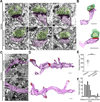


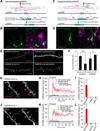
Similar articles
-
Genome editing-based approaches for imaging protein localization and dynamics in the mammalian brain.Neurosci Res. 2020 Jan;150:2-7. doi: 10.1016/j.neures.2019.04.007. Epub 2019 Apr 26. Neurosci Res. 2020. PMID: 31034861 Review.
-
Single-cell endogenous protein labeling via CRISPR-Cas9-mediated genome editing in the mouse brain.Anat Sci Int. 2025 Jul 14. doi: 10.1007/s12565-025-00866-x. Online ahead of print. Anat Sci Int. 2025. PMID: 40658346
-
Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain.Neuron. 2017 Nov 15;96(4):755-768.e5. doi: 10.1016/j.neuron.2017.10.004. Epub 2017 Oct 19. Neuron. 2017. PMID: 29056297 Free PMC article.
-
Optimized CRISPR/Cas9-mediated in vivo genome engineering applicable to monitoring dynamics of endogenous proteins in the mouse neural tissues.Sci Rep. 2019 Aug 5;9(1):11309. doi: 10.1038/s41598-019-47721-4. Sci Rep. 2019. PMID: 31383899 Free PMC article.
-
The application of genome editing in studying hearing loss.Hear Res. 2015 Sep;327:102-8. doi: 10.1016/j.heares.2015.04.016. Epub 2015 May 15. Hear Res. 2015. PMID: 25987504 Free PMC article. Review.
Cited by
-
Dendritic Spine Plasticity: Function and Mechanisms.Front Synaptic Neurosci. 2020 Aug 28;12:36. doi: 10.3389/fnsyn.2020.00036. eCollection 2020. Front Synaptic Neurosci. 2020. PMID: 32982715 Free PMC article.
-
Advanced Techniques Using In Vivo Electroporation to Study the Molecular Mechanisms of Cerebral Development Disorders.Int J Mol Sci. 2023 Sep 15;24(18):14128. doi: 10.3390/ijms241814128. Int J Mol Sci. 2023. PMID: 37762431 Free PMC article. Review.
-
Genetically Engineering the Nervous System with CRISPR-Cas.eNeuro. 2020 Mar 25;7(2):ENEURO.0419-19.2020. doi: 10.1523/ENEURO.0419-19.2020. Print 2020 Mar/Apr. eNeuro. 2020. PMID: 32098761 Free PMC article. Review.
-
Spatiotemporal Insights Into RNA-Organelle Interactions in Neurons.Front Cell Dev Biol. 2021 Jun 10;9:663367. doi: 10.3389/fcell.2021.663367. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34178987 Free PMC article. Review.
-
The pain target NaV1.7 is expressed late during human iPS cell differentiation into sensory neurons as determined in high-resolution imaging.Pflugers Arch. 2024 Jun;476(6):975-992. doi: 10.1007/s00424-024-02945-w. Epub 2024 Mar 27. Pflugers Arch. 2024. PMID: 38538988 Free PMC article.
References
-
- Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. - PubMed
-
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. - PubMed
-
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

