Translin and Trax differentially regulate telomere-associated transcript homeostasis
- PMID: 27183912
- PMCID: PMC5085120
- DOI: 10.18632/oncotarget.9278
Translin and Trax differentially regulate telomere-associated transcript homeostasis
Abstract
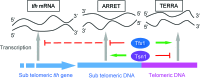
Translin and Trax proteins are highly conserved nucleic acid binding proteins that have been implicated in RNA regulation in a range of biological processes including tRNA processing, RNA interference, microRNA degradation during oncogenesis, spermatogenesis and neuronal regulation. Here, we explore the function of this paralogue pair of proteins in the fission yeast. Using transcript analysis we demonstrate a reciprocal mechanism for control of telomere-associated transcripts. Mutation of tfx1+ (Trax) elevates transcript levels from silenced sub-telomeric regions of the genome, but not other silenced regions, such as the peri-centromeric heterochromatin. In the case of some sub-telomeric transcripts, but not all, this elevation is dependent on the Trax paralogue, Tsn1 (Translin). In a reciprocal fashion, Tsn1 (Translin) serves to repress levels of transcripts (TERRAs) from the telomeric repeats, whereas Tfx1 serves to maintain these elevated levels. This reveals a novel mechanism for the regulation of telomeric transcripts. We extend this to demonstrate that human Translin and Trax also control telomere-associated transcript levels in human cells in a telomere-specific fashion.
Keywords: C3PO; Chromosome Section; TERRA; Translin; Trax; telomeres.
Conflict of interest statement
There are no known conflicts of interest associated with this work.
Figures

Similar articles
-
Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX.Nucleic Acids Res. 2005 Jul 25;33(13):4128-39. doi: 10.1093/nar/gki727. Print 2005. Nucleic Acids Res. 2005. PMID: 16043634 Free PMC article.
-
Identification of proteins that form specific complexes with the highly conserved protein Translin in Schizosaccharomyces pombe.Biochim Biophys Acta. 2014 Apr;1844(4):767-77. doi: 10.1016/j.bbapap.2013.12.016. Epub 2013 Dec 29. Biochim Biophys Acta. 2014. PMID: 24382491
-
Functional characterisation of the Schizosaccharomyces pombe homologue of the leukaemia-associated translocation breakpoint binding protein translin and its binding partner, TRAX.Biochim Biophys Acta. 2008 Feb;1783(2):203-13. doi: 10.1016/j.bbamcr.2007.10.014. Epub 2007 Nov 7. Biochim Biophys Acta. 2008. PMID: 18062930
-
Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore.Biochem J. 2010 Jul 15;429(2):225-34. doi: 10.1042/BJ20100273. Biochem J. 2010. PMID: 20578993 Review.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
-
Structural insights into Drosophila-C3PO complex assembly and 'Dynamic Side Port' model in substrate entry and release.Nucleic Acids Res. 2018 Sep 19;46(16):8590-8604. doi: 10.1093/nar/gky465. Nucleic Acids Res. 2018. PMID: 29860349 Free PMC article.
-
The TRAX, DISC1, and GSK3 complex in mental disorders and therapeutic interventions.J Biomed Sci. 2018 Oct 4;25(1):71. doi: 10.1186/s12929-018-0473-x. J Biomed Sci. 2018. PMID: 30285728 Free PMC article. Review.
-
Translin facilitates RNA polymerase II dissociation and suppresses genome instability during RNase H2- and Dicer-deficiency.PLoS Genet. 2022 Jun 17;18(6):e1010267. doi: 10.1371/journal.pgen.1010267. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35714159 Free PMC article.
-
Human germ/stem cell-specific gene TEX19 influences cancer cell proliferation and cancer prognosis.Mol Cancer. 2017 Apr 26;16(1):84. doi: 10.1186/s12943-017-0653-4. Mol Cancer. 2017. PMID: 28446200 Free PMC article.
-
Subtelomeric Transcription and its Regulation.J Mol Biol. 2020 Jul 10;432(15):4199-4219. doi: 10.1016/j.jmb.2020.01.026. Epub 2020 Feb 6. J Mol Biol. 2020. PMID: 32035903 Free PMC article. Review.
References
-
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–485. - PubMed
-
- Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–269. - PubMed
-
- Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40:504–515. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

