CRISPRDetect: A flexible algorithm to define CRISPR arrays
- PMID: 27184979
- PMCID: PMC4869251
- DOI: 10.1186/s12864-016-2627-0
CRISPRDetect: A flexible algorithm to define CRISPR arrays
Abstract
Background: CRISPR (clustered regularly interspaced short palindromic repeats) RNAs provide the specificity for noncoding RNA-guided adaptive immune defence systems in prokaryotes. CRISPR arrays consist of repeat sequences separated by specific spacer sequences. CRISPR arrays have previously been identified in a large proportion of prokaryotic genomes. However, currently available detection algorithms do not utilise recently discovered features regarding CRISPR loci.
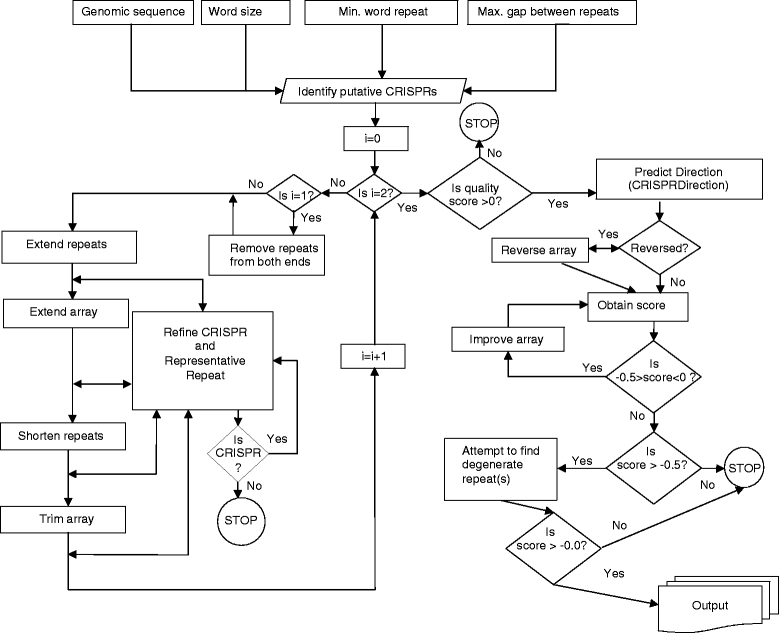
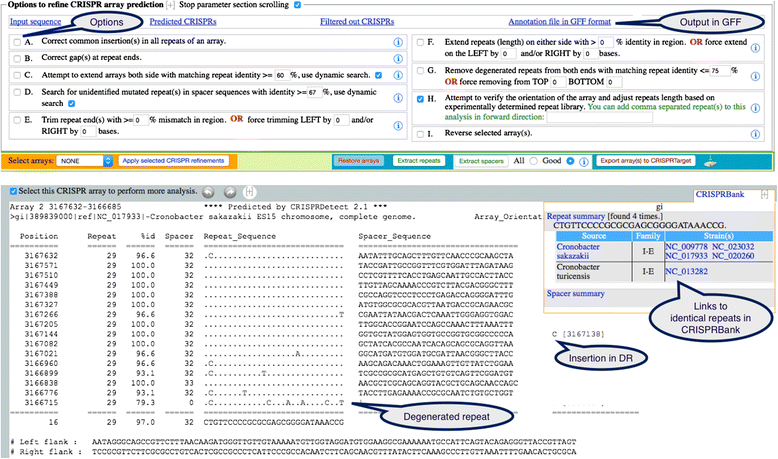
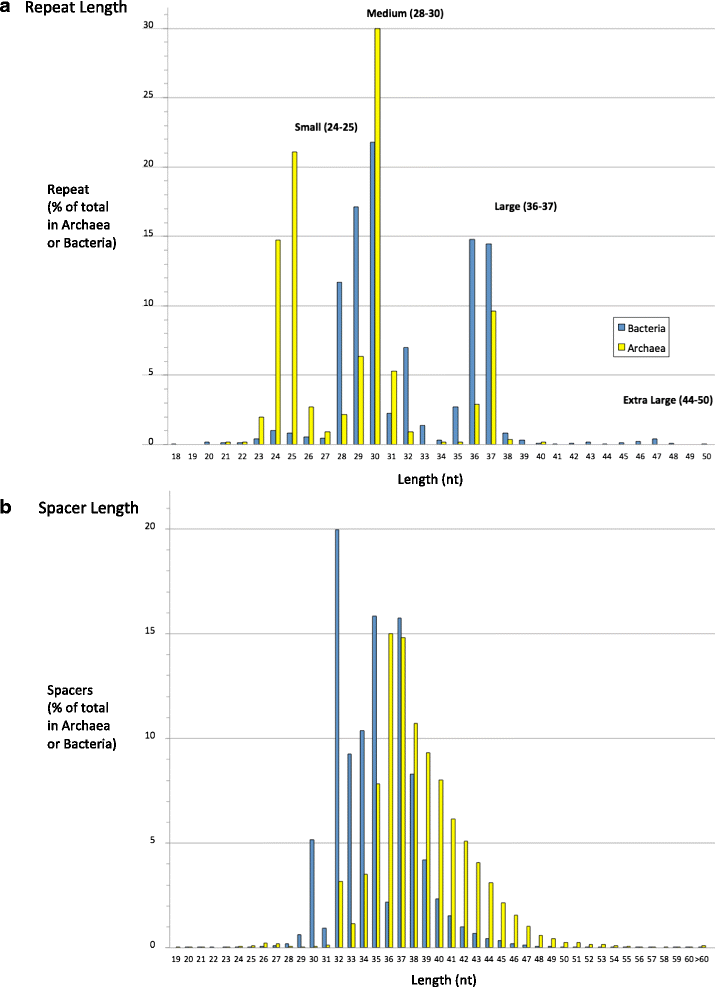
Results: We have developed a new approach to automatically detect, predict and interactively refine CRISPR arrays. It is available as a web program and command line from bioanalysis.otago.ac.nz/CRISPRDetect. CRISPRDetect discovers putative arrays, extends the array by detecting additional variant repeats, corrects the direction of arrays, refines the repeat/spacer boundaries, and annotates different types of sequence variations (e.g. insertion/deletion) in near identical repeats. Due to these features, CRISPRDetect has significant advantages when compared to existing identification tools. As well as further support for small medium and large repeats, CRISPRDetect identified a class of arrays with 'extra-large' repeats in bacteria (repeats 44-50 nt). The CRISPRDetect output is integrated with other analysis tools. Notably, the predicted spacers can be directly utilised by CRISPRTarget to predict targets.
Conclusion: CRISPRDetect enables more accurate detection of arrays and spacers and its gff output is suitable for inclusion in genome annotation pipelines and visualisation. It has been used to analyse all complete bacterial and archaeal reference genomes.
Keywords: Bioinformatics; CRISPR; Cas; Horizontal gene transfer; Phage resistance; Plasmids; Repeat elements; Small RNA targets; crRNA.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

