Integrated metabolomic and transcriptome analyses reveal finishing forage affects metabolic pathways related to beef quality and animal welfare
- PMID: 27185157
- PMCID: PMC4869019
- DOI: 10.1038/srep25948
Integrated metabolomic and transcriptome analyses reveal finishing forage affects metabolic pathways related to beef quality and animal welfare
Abstract
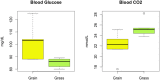
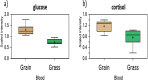
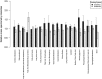
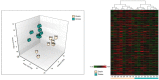
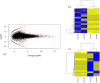
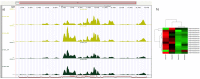
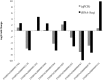
Beef represents a major dietary component and source of protein in many countries. With an increasing demand for beef, the industry is currently undergoing changes towards naturally produced beef. However, the true differences between the feeding systems, especially the biochemical and nutritional aspects, are still unclear. Using transcriptome and metabolome profiles, we identified biological pathways related to the differences between grass- and grain-fed Angus steers. In the latissimus dorsi muscle, we have recognized 241 differentially expressed genes (FDR < 0.1). The metabolome examinations of muscle and blood revealed 163 and 179 altered compounds in each tissue (P < 0.05), respectively. Accordingly, alterations in glucose metabolism, divergences in free fatty acids and carnitine conjugated lipid levels, and altered β-oxidation have been observed. The anti-inflammatory n3 polyunsaturated fatty acids are enriched in grass finished beef, while higher levels of n6 PUFAs in grain finished animals may promote inflammation and oxidative stress. Furthermore, grass-fed animals produce tender beef with lower total fat and a higher omega3/omega6 ratio than grain-fed ones, which could potentially benefit consumer health. Most importantly, blood cortisol levels strongly indicate that grass-fed animals may experience less stress than the grain-fed individuals. These results will provide deeper insights into the merits and mechanisms of muscle development.
Figures



References
-
- Realini C. E., Duckett S. K. & Windham W. R. Effect of vitamin C addition to ground beef from grass-fed or grain-fed sources on color and lipid stability, and prediction of fatty acid composition by near-infrared reflectance analysis. Meat Sci 68, 35–43, doi: 10.1016/j.meatsci.2004.02.002 (2004). - DOI - PubMed
-
- Xue H., Mainville D., You W. & Nayga R. M. Jr Consumer preferences and willingness to pay for grass-fed beef: Empirical evidence from in-store experiments. Food Quality and Preference 21, 857–866, doi: 10.1016/j.foodqual.2010.05.004 (2010). - DOI
-
- Roosevelt M. The grass-fed revolution. Beef raised wholly on pasture, rather than grain-fed in feedlots, may be better for your health–and for the planet. Time 167, 76–78 (2006). - PubMed
-
- Roberts J. L. The prevalence and economic significance of liver disorders and contamination in grain-fed and grass-fed cattle. Aust Vet J 59, 129–132 (1982). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

