Homozygosity for a haplotype in the HBG2-OR51B4 region is exclusive to Arab-Indian haplotype sickle cell anemia
- PMID: 27185208
- PMCID: PMC4874527
- DOI: 10.1002/ajh.24368
Homozygosity for a haplotype in the HBG2-OR51B4 region is exclusive to Arab-Indian haplotype sickle cell anemia
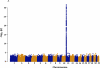
Figures

References
-
- Perrine RP, Brown MJ, Clegg JB, et al. Benign sickle-cell anaemia. Lancet. 1972;2:1163–1167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
