Asterless is required for centriole length control and sperm development
- PMID: 27185836
- PMCID: PMC4878089
- DOI: 10.1083/jcb.201501120
Asterless is required for centriole length control and sperm development
Abstract
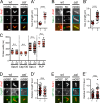
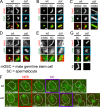
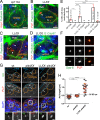
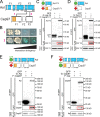
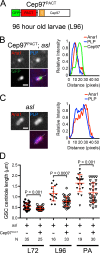
Centrioles are the foundation of two organelles, centrosomes and cilia. Centriole numbers and functions are tightly controlled, and mutations in centriole proteins are linked to a variety of diseases, including microcephaly. Loss of the centriole protein Asterless (Asl), the Drosophila melanogaster orthologue of Cep152, prevents centriole duplication, which has limited the study of its nonduplication functions. Here, we identify populations of cells with Asl-free centrioles in developing Drosophila tissues, allowing us to assess its duplication-independent function. We show a role for Asl in controlling centriole length in germline and somatic tissue, functioning via the centriole protein Cep97. We also find that Asl is not essential for pericentriolar material recruitment or centrosome function in organizing mitotic spindles. Lastly, we show that Asl is required for proper basal body function and spermatid axoneme formation. Insights into the role of Asl/Cep152 beyond centriole duplication could help shed light on how Cep152 mutations lead to the development of microcephaly.
Figures




Similar articles
-
Asterless is a scaffold for the onset of centriole assembly.Nature. 2010 Oct 7;467(7316):714-8. doi: 10.1038/nature09445. Epub 2010 Sep 19. Nature. 2010. PMID: 20852615
-
Conserved molecular interactions in centriole-to-centrosome conversion.Nat Cell Biol. 2016 Jan;18(1):87-99. doi: 10.1038/ncb3274. Epub 2015 Nov 23. Nat Cell Biol. 2016. PMID: 26595382 Free PMC article.
-
Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication.Genetics. 2008 Dec;180(4):2081-94. doi: 10.1534/genetics.108.095141. Epub 2008 Oct 14. Genetics. 2008. PMID: 18854586 Free PMC article.
-
Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification.Cell Mol Life Sci. 2013 Mar;70(6):1021-34. doi: 10.1007/s00018-012-1102-6. Epub 2012 Aug 15. Cell Mol Life Sci. 2013. PMID: 22892665 Free PMC article. Review.
-
The Centrioles, Centrosomes, Basal Bodies, and Cilia of Drosophila melanogaster.Genetics. 2017 May;206(1):33-53. doi: 10.1534/genetics.116.198168. Genetics. 2017. PMID: 28476861 Free PMC article. Review.
Cited by
-
Bridging centrioles and PCM in proper space and time.Essays Biochem. 2018 Dec 7;62(6):793-801. doi: 10.1042/EBC20180036. Print 2018 Dec 7. Essays Biochem. 2018. PMID: 30429283 Free PMC article. Review.
-
Regulation of organelle size and organization during development.Semin Cell Dev Biol. 2023 Jan 15;133:53-64. doi: 10.1016/j.semcdb.2022.02.002. Epub 2022 Feb 8. Semin Cell Dev Biol. 2023. PMID: 35148938 Free PMC article. Review.
-
CCDC15 localizes to the centriole inner scaffold and controls centriole length and integrity.J Cell Biol. 2023 Dec 4;222(12):e202305009. doi: 10.1083/jcb.202305009. Epub 2023 Nov 7. J Cell Biol. 2023. PMID: 37934472 Free PMC article.
-
Single-cell transcriptomic atlas of the human testis across the reproductive lifespan.Nat Aging. 2025 Apr;5(4):658-674. doi: 10.1038/s43587-025-00824-2. Epub 2025 Mar 3. Nat Aging. 2025. PMID: 40033047 Free PMC article.
-
SAPs as a new model to probe the pathway of centriole and centrosome assembly.Biochem Soc Trans. 2021 Jun 30;49(3):1233-1240. doi: 10.1042/BST20200833. Biochem Soc Trans. 2021. PMID: 33960367 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

