Brg1 Enables Rapid Growth of the Early Embryo by Suppressing Genes That Regulate Apoptosis and Cell Growth Arrest
- PMID: 27185875
- PMCID: PMC4946428
- DOI: 10.1128/MCB.01101-15
Brg1 Enables Rapid Growth of the Early Embryo by Suppressing Genes That Regulate Apoptosis and Cell Growth Arrest
Abstract
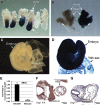
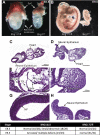
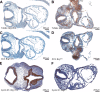
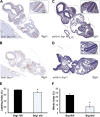
SWI/SNF (switching/sucrose nonfermenting)-dependent chromatin remodeling establishes coordinated gene expression programs during development, yet important functional details remain to be elucidated. We show that the Brg1 (Brahma-related gene 1; Smarca4) ATPase is globally expressed at high levels during postimplantation development and its conditional ablation, beginning at gastrulation, results in increased apoptosis, growth retardation, and, ultimately, embryonic death. Global gene expression analysis revealed that genes upregulated in Rosa26CreERT2; Brg1(flox/flox) embryos (here referred to as Brg1(d/d) embryos to describe embryos with deletion of the Brg1(flox/flox) alleles) negatively regulate cell cycle progression and cell growth. In addition, the p53 (Trp53) protein, which is virtually undetectable in early wild-type embryos, accumulated in the Brg1(d/d) embryos and activated the p53-dependent pathways. Using P19 cells, we show that Brg1 and CHD4 (chromodomain helicase DNA binding protein 4) coordinate to control target gene expression. Both proteins physically interact and show a substantial overlap of binding sites at chromatin-accessible regions adjacent to genes differentially expressed in the Brg1(d/d) embryos. Specifically, Brg1 deficiency results in reduced levels of the repressive histone H3 lysine K27 trimethylation (H3K27me3) histone mark and an increase in the amount of open chromatin at the regulatory region of the p53 and p21 (Cdkn1a) genes. These results provide insights into the mechanisms by which Brg1 functions, which is in part via the p53 program, to constrain gene expression and facilitate rapid embryonic growth.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures










Similar articles
-
The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells.Oncogene. 2009 Jul 9;28(27):2492-501. doi: 10.1038/onc.2009.121. Epub 2009 May 18. Oncogene. 2009. PMID: 19448667 Free PMC article.
-
Endothelial Chromatin-Remodeling Enzymes Regulate the Production of Critical ECM Components During Murine Lung Development.Arterioscler Thromb Vasc Biol. 2024 Aug;44(8):1784-1798. doi: 10.1161/ATVBAHA.124.320881. Epub 2024 Jun 13. Arterioscler Thromb Vasc Biol. 2024. PMID: 38868942 Free PMC article.
-
A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1.Cancer Res. 2013 Sep 1;73(17):5508-18. doi: 10.1158/0008-5472.CAN-12-4593. Epub 2013 Jul 19. Cancer Res. 2013. PMID: 23872584
-
The Role of BRG1 in Antioxidant and Redox Signaling.Oxid Med Cell Longev. 2020 Sep 14;2020:6095673. doi: 10.1155/2020/6095673. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33014273 Free PMC article. Review.
-
Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer.Epigenetics. 2008 Mar-Apr;3(2):64-8. doi: 10.4161/epi.3.2.6153. Epub 2008 Apr 17. Epigenetics. 2008. PMID: 18437052 Review.
Cited by
-
BRG1: Promoter or Suppressor of Cancer? The Outcome of BRG1's Interaction with Specific Cellular Pathways.Int J Mol Sci. 2023 Feb 2;24(3):2869. doi: 10.3390/ijms24032869. Int J Mol Sci. 2023. PMID: 36769189 Free PMC article. Review.
-
ARID1A, a component of SWI/SNF chromatin remodeling complexes, is required for porcine embryo development.Mol Reprod Dev. 2017 Dec;84(12):1250-1256. doi: 10.1002/mrd.22924. Epub 2017 Nov 26. Mol Reprod Dev. 2017. PMID: 29178559 Free PMC article.
-
SWI/SNF chromatin remodeling subunit Smarca4/BRG1 is essential for female fertility†.Biol Reprod. 2023 Feb 13;108(2):279-291. doi: 10.1093/biolre/ioac209. Biol Reprod. 2023. PMID: 36440965 Free PMC article.
-
Impact of carbamazepine on SMARCA4 (BRG1) expression in colorectal cancer: modulation by KRAS mutation status.Invest New Drugs. 2024 Apr;42(2):229-239. doi: 10.1007/s10637-024-01418-2. Epub 2024 Mar 6. Invest New Drugs. 2024. PMID: 38446332 Free PMC article.
-
BRG1 Activates Proliferation and Transcription of Cell Cycle-Dependent Genes in Breast Cancer Cells.Cancers (Basel). 2020 Feb 4;12(2):349. doi: 10.3390/cancers12020349. Cancers (Basel). 2020. PMID: 32033115 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
