incaRNAfbinv: a web server for the fragment-based design of RNA sequences
- PMID: 27185893
- PMCID: PMC5741205
- DOI: 10.1093/nar/gkw440
incaRNAfbinv: a web server for the fragment-based design of RNA sequences
Abstract
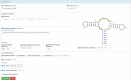
In recent years, new methods for computational RNA design have been developed and applied to various problems in synthetic biology and nanotechnology. Lately, there is considerable interest in incorporating essential biological information when solving the inverse RNA folding problem. Correspondingly, RNAfbinv aims at including biologically meaningful constraints and is the only program to-date that performs a fragment-based design of RNA sequences. In doing so it allows the design of sequences that do not necessarily exactly fold into the target, as long as the overall coarse-grained tree graph shape is preserved. Augmented by the weighted sampling algorithm of incaRNAtion, our web server called incaRNAfbinv implements the method devised in RNAfbinv and offers an interactive environment for the inverse folding of RNA using a fragment-based design approach. It takes as input: a target RNA secondary structure; optional sequence and motif constraints; optional target minimum free energy, neutrality and GC content. In addition to the design of synthetic regulatory sequences, it can be used as a pre-processing step for the detection of novel natural occurring RNAs. The two complementary methodologies RNAfbinv and incaRNAtion are merged together and fully implemented in our web server incaRNAfbinv, available at http://www.cs.bgu.ac.il/incaRNAfbinv.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Design of RNAs: comparing programs for inverse RNA folding.Brief Bioinform. 2018 Mar 1;19(2):350-358. doi: 10.1093/bib/bbw120. Brief Bioinform. 2018. PMID: 28049135 Free PMC article. Review.
-
RNAfbinv: an interactive Java application for fragment-based design of RNA sequences.Bioinformatics. 2013 Nov 15;29(22):2938-40. doi: 10.1093/bioinformatics/btt494. Epub 2013 Aug 23. Bioinformatics. 2013. PMID: 23975763
-
incaRNAfbinv 2.0: a webserver and software with motif control for fragment-based design of RNAs.Bioinformatics. 2020 May 1;36(9):2920-2922. doi: 10.1093/bioinformatics/btaa039. Bioinformatics. 2020. PMID: 31971575
-
Rtools: a web server for various secondary structural analyses on single RNA sequences.Nucleic Acids Res. 2016 Jul 8;44(W1):W302-7. doi: 10.1093/nar/gkw337. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131356 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
Partial RNA design.Bioinformatics. 2024 Jun 28;40(Suppl 1):i437-i445. doi: 10.1093/bioinformatics/btae222. Bioinformatics. 2024. PMID: 38940170 Free PMC article.
-
MoiRNAiFold: a novel tool for complex in silico RNA design.Nucleic Acids Res. 2021 May 21;49(9):4934-4943. doi: 10.1093/nar/gkab331. Nucleic Acids Res. 2021. PMID: 33956139 Free PMC article.
-
RNA Design Using incaRNAfbinv Demonstrated with the Identification of Functional RNA Motifs in Hepatitis Delta Virus.Methods Mol Biol. 2025;2847:109-120. doi: 10.1007/978-1-0716-4079-1_7. Methods Mol Biol. 2025. PMID: 39312139
-
Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study.Nucleic Acids Res. 2019 Jan 10;47(1):29-42. doi: 10.1093/nar/gky1118. Nucleic Acids Res. 2019. PMID: 30462314 Free PMC article.
-
Design of RNAs: comparing programs for inverse RNA folding.Brief Bioinform. 2018 Mar 1;19(2):350-358. doi: 10.1093/bib/bbw120. Brief Bioinform. 2018. PMID: 28049135 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

