Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression
- PMID: 27185910
- PMCID: PMC4896695
- DOI: 10.1073/pnas.1600420113
Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression
Abstract
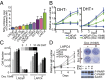
The androgen receptor (AR) plays an essential role in prostate cancer, and suppression of its signaling with androgen deprivation therapy (ADT) has been the mainstay of treatment for metastatic hormone-sensitive prostate cancer for more than 70 y. Chemotherapy has been reserved for metastatic castration-resistant prostate cancer (mCRPC). The Eastern Cooperative Oncology Group-led trial E3805: ChemoHormonal Therapy Versus Androgen Ablation Randomized Trial for Extensive Disease in Prostate Cancer (CHAARTED) showed that the addition of docetaxel to ADT prolonged overall survival compared with ADT alone in patients with metastatic hormone-sensitive prostate cancer. This finding suggests that there is an interaction between AR signaling activity and docetaxel sensitivity. Here we demonstrate that the prostate cancer cell lines LNCaP and LAPC4 display markedly different sensitivity to docetaxel with AR activation, and RNA-seq analysis of these cell lines identified KDM5D (lysine-specific demethylase 5D) encoded on the Y chromosome as a potential mediator of this sensitivity. Knocking down KDM5D expression in LNCaP leads to docetaxel resistance in the presence of dihydrotestosterone. KDM5D physically interacts with AR in the nucleus, and regulates its transcriptional activity by demethylating H3K4me3 active transcriptional marks. Attenuating KDM5D expression dysregulates AR signaling, resulting in docetaxel insensitivity. KDM5D deletion was also observed in the LNCaP-derived CRPC cell line 104R2, which displayed docetaxel insensitivity with AR activation, unlike parental LNCaP. Dataset analysis from the Oncomine database revealed significantly decreased KDM5D expression in CRPC and poorer prognosis with low KDM5D expression. Taking these data together, this work indicates that KDM5D modulates the AR axis and that this is associated with altered docetaxel sensitivity.
Keywords: JARID1D; KDM5D; androgen receptor; docetaxel; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Comment in
-
Re: Resistance to Docetaxel in Prostate Cancer is Associated with Androgen Receptor Activation and Loss of KDM5D Expression.J Urol. 2017 Jan;197(1):154-155. doi: 10.1016/j.juro.2016.10.010. Epub 2016 Oct 8. J Urol. 2017. PMID: 27979512 No abstract available.
Similar articles
-
Increased MYBL2 expression in aggressive hormone-sensitive prostate cancer.Mol Oncol. 2022 Dec;16(22):3994-4010. doi: 10.1002/1878-0261.13314. Epub 2022 Oct 2. Mol Oncol. 2022. PMID: 36087093 Free PMC article.
-
Exploitation of the Androgen Receptor to Overcome Taxane Resistance in Advanced Prostate Cancer.Adv Cancer Res. 2015;127:123-58. doi: 10.1016/bs.acr.2015.03.001. Epub 2015 Mar 29. Adv Cancer Res. 2015. PMID: 26093899 Review.
-
Androgen receptor negatively regulates mitotic checkpoint signaling to induce docetaxel resistance in castration-resistant prostate cancer.Prostate. 2022 Feb;82(2):182-192. doi: 10.1002/pros.24257. Epub 2021 Oct 21. Prostate. 2022. PMID: 34672379 Free PMC article.
-
Interaction between docetaxel resistance and castration resistance in prostate cancer: implications of Twist1, YB-1, and androgen receptor.Prostate. 2013 Sep;73(12):1336-44. doi: 10.1002/pros.22681. Epub 2013 Jun 14. Prostate. 2013. PMID: 23775496
-
Targeting Metastatic Hormone Sensitive Prostate Cancer: Chemohormonal Therapy and New Combinatorial Approaches.J Urol. 2019 May;201(5):876-885. doi: 10.1097/JU.0000000000000117. J Urol. 2019. PMID: 30747897 Review.
Cited by
-
Prostate epithelial genes define therapy-relevant prostate cancer molecular subtype.Prostate Cancer Prostatic Dis. 2021 Dec;24(4):1080-1092. doi: 10.1038/s41391-021-00364-x. Epub 2021 Apr 26. Prostate Cancer Prostatic Dis. 2021. PMID: 33903734 Free PMC article.
-
The role of microRNA-572 in the proliferation and chemotherapeutic treatment of prostate cancer.J Int Med Res. 2021 May;49(5):3000605211014363. doi: 10.1177/03000605211014363. J Int Med Res. 2021. PMID: 34044640 Free PMC article.
-
Myc and ChREBP transcription factors cooperatively regulate normal and neoplastic hepatocyte proliferation in mice.J Biol Chem. 2018 Sep 21;293(38):14740-14757. doi: 10.1074/jbc.RA118.004099. Epub 2018 Aug 7. J Biol Chem. 2018. PMID: 30087120 Free PMC article.
-
Pathogenic and Therapeutic Role of H3K4 Family of Methylases and Demethylases in Cancers.Indian J Clin Biochem. 2019 Apr;34(2):123-132. doi: 10.1007/s12291-019-00828-x. Epub 2019 Apr 3. Indian J Clin Biochem. 2019. PMID: 31092985 Free PMC article. Review.
-
Consequences of Y chromosome microdeletions beyond male infertility.J Assist Reprod Genet. 2019 Jul;36(7):1329-1337. doi: 10.1007/s10815-019-01492-z. Epub 2019 Jun 18. J Assist Reprod Genet. 2019. PMID: 31214882 Free PMC article. Review.
References
-
- Petrylak DP, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351(15):1513–1520. - PubMed
-
- Tannock IF, et al. TAX 327 Investigators Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–1512. - PubMed
-
- James ND, et al. STAMPEDE Investigators Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): Survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163–1177. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

