Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation
- PMID: 27185912
- PMCID: PMC4896708
- DOI: 10.1073/pnas.1519881113
Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation
Abstract
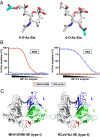
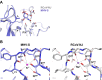
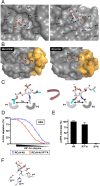
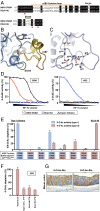
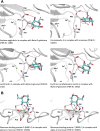
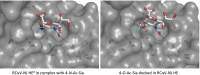
Hemagglutinin-esterases (HEs) are bimodular envelope proteins of orthomyxoviruses, toroviruses, and coronaviruses with a carbohydrate-binding "lectin" domain appended to a receptor-destroying sialate-O-acetylesterase ("esterase"). In concert, these domains facilitate dynamic virion attachment to cell-surface sialoglycans. Most HEs (type I) target 9-O-acetylated sialic acids (9-O-Ac-Sias), but one group of coronaviruses switched to using 4-O-Ac-Sias instead (type II). This specificity shift required quasisynchronous adaptations in the Sia-binding sites of both lectin and esterase domains. Previously, a partially disordered crystal structure of a type II HE revealed how the shift in lectin ligand specificity was achieved. How the switch in esterase substrate specificity was realized remained unresolved, however. Here, we present a complete structure of a type II HE with a receptor analog in the catalytic site and identify the mutations underlying the 9-O- to 4-O-Ac-Sia substrate switch. We show that (i) common principles pertaining to the stereochemistry of protein-carbohydrate interactions were at the core of the transition in lectin ligand and esterase substrate specificity; (ii) in consequence, the switch in O-Ac-Sia specificity could be readily accomplished via convergent intramolecular coevolution with only modest architectural changes in lectin and esterase domains; and (iii) a single, inconspicuous Ala-to-Ser substitution in the catalytic site was key to the emergence of the type II HEs. Our findings provide fundamental insights into how proteins "see" sugars and how this affects protein and virus evolution.
Keywords: coronavirus; crystal structure; hemagglutinin-esterase; sialate-O-acetyl esterase; sialic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases.Proc Natl Acad Sci U S A. 2009 Sep 15;106(37):15897-902. doi: 10.1073/pnas.0904266106. Epub 2009 Aug 31. Proc Natl Acad Sci U S A. 2009. PMID: 19721004 Free PMC article.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
-
The murine coronavirus hemagglutinin-esterase receptor-binding site: a major shift in ligand specificity through modest changes in architecture.PLoS Pathog. 2012 Jan;8(1):e1002492. doi: 10.1371/journal.ppat.1002492. Epub 2012 Jan 26. PLoS Pathog. 2012. PMID: 22291594 Free PMC article.
-
Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25759-25770. doi: 10.1073/pnas.2006299117. Epub 2020 Sep 29. Proc Natl Acad Sci U S A. 2020. PMID: 32994342 Free PMC article.
-
Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses.Glycoconj J. 2006 Feb;23(1-2):59-72. doi: 10.1007/s10719-006-5438-8. Glycoconj J. 2006. PMID: 16575523 Free PMC article. Review.
Cited by
-
Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle.Viruses. 2022 Sep 27;14(10):2125. doi: 10.3390/v14102125. Viruses. 2022. PMID: 36298681 Free PMC article.
-
SDAV, the Rat Coronavirus-How Much Do We Know about It in the Light of Potential Zoonoses.Viruses. 2021 Oct 4;13(10):1995. doi: 10.3390/v13101995. Viruses. 2021. PMID: 34696425 Free PMC article. Review.
-
Structure of the infectious salmon anemia virus receptor complex illustrates a unique binding strategy for attachment.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):E2929-E2936. doi: 10.1073/pnas.1617993114. Epub 2017 Mar 20. Proc Natl Acad Sci U S A. 2017. PMID: 28320973 Free PMC article.
-
Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans.Nat Commun. 2020 Sep 16;11(1):4646. doi: 10.1038/s41467-020-18440-6. Nat Commun. 2020. PMID: 32938911 Free PMC article.
-
Host Receptors of Influenza Viruses and Coronaviruses-Molecular Mechanisms of Recognition.Vaccines (Basel). 2020 Oct 6;8(4):587. doi: 10.3390/vaccines8040587. Vaccines (Basel). 2020. PMID: 33036202 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

