BPTF transduces MITF-driven prosurvival signals in melanoma cells
- PMID: 27185926
- PMCID: PMC4896689
- DOI: 10.1073/pnas.1606027113
BPTF transduces MITF-driven prosurvival signals in melanoma cells
Abstract
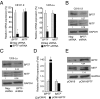
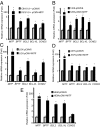
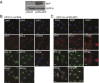
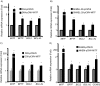
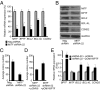
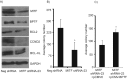
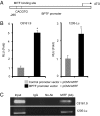
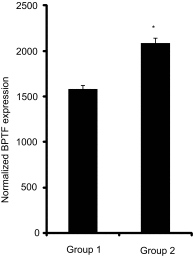
Microphthalmia-associated transcription factor (MITF) plays a critical and complex role in melanocyte transformation. Although several downstream targets of MITF action have been identified, the precise mechanisms by which MITF promotes melanocytic tumor progression are incompletely understood. Recent studies identified an oncogenic role for the bromodomain plant homeodomain finger transcription factor (BPTF) gene in melanoma progression, in part through activation of BCL2, a canonical target of MITF signaling. Analysis of the BPTF promoter identified a putative MITF-binding site, suggesting that MITF may regulate BPTF expression. Overexpression of MITF resulted in up-regulation of BPTF in a panel of melanoma and melanocyte cell lines. shRNA-mediated down-regulation of MITF in melanoma cells was accompanied by down-regulation of BPTF and BPTF-regulated genes (including BCL2) and resulted in reduced proliferative capacity of melanoma cells. The suppression of cell growth mediated by MITF silencing was rescued by overexpression of BPTF cDNA. Binding of MITF to the BPTF promoter was demonstrated using ChIP analysis. MITF overexpression resulted in direct transcriptional activation of BPTF, as evidenced by increased luciferase activity driven by the BPTF promoter. These results indicate that BPTF transduces key prosurvival signals driven by MITF, further supporting its important role in promoting melanoma cell survival and progression.
Keywords: melanoma; oncogenes; signaling cascade.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
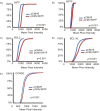
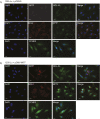
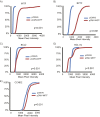
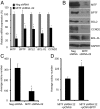
References
-
- Yavuzer U, et al. The Microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10(1):123–134. - PubMed
-
- Levy C, Khaled M, Fisher DE. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12(9):406–414. - PubMed
-
- Widlund HR, Fisher DE. Microphthalamia-associated transcription factor: A critical regulator of pigment cell development and survival. Oncogene. 2003;22(20):3035–3041. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

