Open chromatin reveals the functional maize genome
- PMID: 27185945
- PMCID: PMC4896728
- DOI: 10.1073/pnas.1525244113
Open chromatin reveals the functional maize genome
Abstract
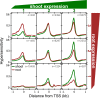
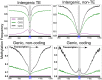
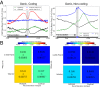
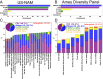
Cellular processes mediated through nuclear DNA must contend with chromatin. Chromatin structural assays can efficiently integrate information across diverse regulatory elements, revealing the functional noncoding genome. In this study, we use a differential nuclease sensitivity assay based on micrococcal nuclease (MNase) digestion to discover open chromatin regions in the maize genome. We find that maize MNase-hypersensitive (MNase HS) regions localize around active genes and within recombination hotspots, focusing biased gene conversion at their flanks. Although MNase HS regions map to less than 1% of the genome, they consistently explain a remarkably large amount (∼40%) of heritable phenotypic variance in diverse complex traits. MNase HS regions are therefore on par with coding sequences as annotations that demarcate the functional parts of the maize genome. These results imply that less than 3% of the maize genome (coding and MNase HS regions) may give rise to the overwhelming majority of phenotypic variation, greatly narrowing the scope of the functional genome.
Keywords: biased gene conversion; chromatin; maize; recombination; variance partitioning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

