Structural characterization of the ternary complex that mediates termination of NF-κB signaling by IκBα
- PMID: 27185953
- PMCID: PMC4896678
- DOI: 10.1073/pnas.1603488113
Structural characterization of the ternary complex that mediates termination of NF-κB signaling by IκBα
Abstract
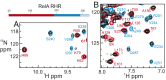
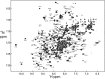
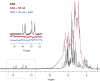
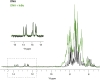
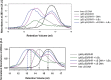
The transcription factor NF-κB is used in many systems for the transduction of extracellular signals into the expression of signal-responsive genes. Published structural data explain the activation of NF-κB through degradation of its dedicated inhibitor IκBα, but the mechanism by which NF-κB-mediated signaling is turned off by its removal from the DNA in the presence of newly synthesized IκBα (termed stripping) is unknown. Previous kinetic studies showed that IκBα accelerates NF-κB dissociation from DNA, and a transient ternary complex between NF-κB, its cognate DNA sequence, and IκBα was observed. Here we structurally characterize the >100-kDa ternary complex by NMR and negative stain EM and show a modeled structure that is consistent with the measurements. These data provide a structural basis for previously unidentified insights into the molecular mechanism of stripping.
Keywords: NMR; negative stain electron microscopy; protein–DNA complex; protein–protein complex; transcriptional activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





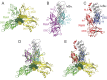

References
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298(5596):1241–1245. - PubMed
-
- Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391(6665):410–413. - PubMed
-
- Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95(6):749–758. - PubMed
-
- Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95(6):759–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

