Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways
- PMID: 27186411
- PMCID: PMC4859668
Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways
Abstract
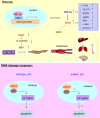
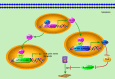
DEAD-box RNA helicase 3 (DDX3) is a highly conserved family member of DEAD-box protein, which is a cluster of ATP-dependent and the largest family of RNA helicase. DEAD-box family is characterized by the regulation of ATPase and helicase activities, the modulation of RNA metabolism, and the actors of RNA binding proteins or molecular chaperones to interact with other proteins or RNA. For DDX3, it exerts its multifaceted roles in viral manipulation, stress response, hypoxia, radiation response and apoptosis, and is closely related to cancer development and progression. DDX3 has dual roles in different cancer types and can act as either an oncogene or tumor suppressor gene during cancer progression. In the present review, we mainly provide an overview of current knowledge on dual roles of DDX3 in various types of cancer, including breast cancer, lung cancer, colorectal cancer, hepatocellular carcinoma, oral squamous cell carcinoma, Ewing sarcoma, glioblastoma multiforme and gallbladder carcinoma, and illustrate the regulatory mechanisms for leading these two controversial biological effects. Furthermore, we summarize the essential signaling pathways that DDX3 participated, especially the Wnt/β-catenin signaling and EMT related signaling (TGF-β, Notch, Hedgehog pathways), which are crucial to DDX3 mediated cancer metastasis process. Thoroughly exploring the dual roles of DDX3 in cancer development and the essential signaling pathways it involved, it will help us open new perspectives to develop novel promising targets to elevate therapeutic effects and facilitate the "Personalized medicine" or "Precision medicine" to come into clinic.
Keywords: DDX3; EMT related pathway; Wnt/β-catenin pathway; cancer; oncogene; tumor suppressor gene.
Figures

Similar articles
-
A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review).Oncol Rep. 2018 Mar;39(3):883-892. doi: 10.3892/or.2018.6203. Epub 2018 Jan 9. Oncol Rep. 2018. PMID: 29328432 Review.
-
Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection.Front Genet. 2014 Dec 5;5:423. doi: 10.3389/fgene.2014.00423. eCollection 2014. Front Genet. 2014. PMID: 25538732 Free PMC article. Review.
-
The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism.Wiley Interdiscip Rev RNA. 2013 Jul-Aug;4(4):369-85. doi: 10.1002/wrna.1165. Epub 2013 Apr 18. Wiley Interdiscip Rev RNA. 2013. PMID: 23606618 Review.
-
The DEAD-Box RNA Helicase DDX3 Interacts with m6A RNA Demethylase ALKBH5.Stem Cells Int. 2017;2017:8596135. doi: 10.1155/2017/8596135. Epub 2017 Nov 23. Stem Cells Int. 2017. PMID: 29333169 Free PMC article.
-
DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential.Eur J Immunol. 2010 Apr;40(4):940-8. doi: 10.1002/eji.200940203. Eur J Immunol. 2010. PMID: 20127681
Cited by
-
The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.Nat Commun. 2019 Jul 12;10(1):3085. doi: 10.1038/s41467-019-11083-2. Nat Commun. 2019. PMID: 31300642 Free PMC article.
-
DDX3 binding with CK1ε was closely related to motor neuron degeneration of ALS by affecting neurite outgrowth.Am J Transl Res. 2017 Oct 15;9(10):4627-4639. eCollection 2017. Am J Transl Res. 2017. PMID: 29118923 Free PMC article.
-
Novel alternative ribonucleotide excision repair pathways in human cells by DDX3X and specialized DNA polymerases.Nucleic Acids Res. 2020 Nov 18;48(20):11551-11565. doi: 10.1093/nar/gkaa948. Nucleic Acids Res. 2020. PMID: 33137198 Free PMC article.
-
Targeting RNA helicase DDX3 in stem cell maintenance and teratoma formation.Genes Cancer. 2019 Feb;10(1-2):11-20. doi: 10.18632/genesandcancer.187. Genes Cancer. 2019. PMID: 30899416 Free PMC article.
-
DDX3X and Stress Granules: Emerging Players in Cancer and Drug Resistance.Cancers (Basel). 2024 Mar 12;16(6):1131. doi: 10.3390/cancers16061131. Cancers (Basel). 2024. PMID: 38539466 Free PMC article. Review.
References
-
- Linder P, Fuller-Pace F. Happy birthday: 25 years of DEAD-box proteins. Methods Mol Biol. 2015;1259:17–33. - PubMed
-
- Andreou AZ, Klostermeier D. Fluorescence methods in the investigation of the DEAD-box helicase mechanism. EXS. 2014;105:161–92. - PubMed
-
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
