Nested Association Mapping of Stem Rust Resistance in Wheat Using Genotyping by Sequencing
- PMID: 27186883
- PMCID: PMC4870046
- DOI: 10.1371/journal.pone.0155760
Nested Association Mapping of Stem Rust Resistance in Wheat Using Genotyping by Sequencing
Abstract
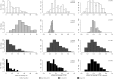
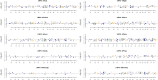
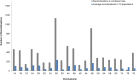
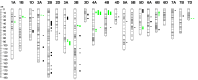
We combined the recently developed genotyping by sequencing (GBS) method with joint mapping (also known as nested association mapping) to dissect and understand the genetic architecture controlling stem rust resistance in wheat (Triticum aestivum). Ten stem rust resistant wheat varieties were crossed to the susceptible line LMPG-6 to generate F6 recombinant inbred lines. The recombinant inbred line populations were phenotyped in Kenya, South Africa, and St. Paul, Minnesota, USA. By joint mapping of the 10 populations, we identified 59 minor and medium-effect QTL (explained phenotypic variance range of 1% - 20%) on 20 chromosomes that contributed towards adult plant resistance to North American Pgt races as well as the highly virulent Ug99 race group. Fifteen of the 59 QTL were detected in multiple environments. No epistatic relationship was detected among the QTL. While these numerous small- to medium-effect QTL are shared among the families, the founder parents were found to have different allelic effects for the QTL. Fourteen QTL identified by joint mapping were also detected in single-population mapping. As these QTL were mapped using SNP markers with known locations on the physical chromosomes, the genomic regions identified with QTL could be explored more in depth to discover candidate genes for stem rust resistance. The use of GBS-derived de novo SNPs in mapping resistance to stem rust shown in this study could be used as a model to conduct similar marker-trait association studies in other plant species.
Conflict of interest statement
Figures

References
-
- Hodson DP, Nazari K, Park RF, Hansen J, Lassen P, Arista J, et al. Putting Ug99 on the map: an update on current and future monitoring2011.
-
- Roelfs AP. Epidemiology in North America In 'The Cereal Rusts'. Vol. II Bushnell WR, Roelfs AP, editors. Orlando, Florida, USA: Academic Press; 1985.
-
- McIntosh RA, Wellings CR, Park RF. Wheat rusts: an atlas of resistance genes. Victoria, Australia: CSIRO Publications; 1995.
-
- Jin Y, Szabo LJ, Rouse MN, FT G., Pretorius ZA, Wanyera R, et al. Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Disease. 2009;93:367–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

