Phenome-Wide Association Study for Alcohol and Nicotine Risk Alleles in 26394 Women
- PMID: 27187070
- PMCID: PMC5026736
- DOI: 10.1038/npp.2016.72
Phenome-Wide Association Study for Alcohol and Nicotine Risk Alleles in 26394 Women
Abstract
To identify novel traits associated with alleles known to predispose to alcohol and nicotine use, we conducted a phenome-wide association study (PheWAS) in a large multi-population cohort. We investigated 7688 African-Americans, 1133 Asian-Americans, 14 081 European-Americans, and 3492 Hispanic-Americans from the Women's Health Initiative, analyzing alleles at the CHRNA3-CHRNA5 locus, ADH1B, and ALDH2 with respect to phenotypic traits related to anthropometric characteristics, dietary habits, social status, psychological traits, reproductive history, health conditions, and nicotine/alcohol use. In ADH1B trans-population meta-analysis and population-specific analysis, we replicated prior associations with drinking behaviors and identified multiple novel phenome-wide significant and suggestive findings related to psychological traits, socioeconomic status, vascular/metabolic conditions, and reproductive health. We then applied Bayesian network learning algorithms to provide insight into the causative relationships of the novel ADH1B associations: ADH1B appears to affect phenotypic traits via both alcohol-mediated and alcohol-independent effects. In an independent sample of 2379 subjects, we also replicated the novel ADH1B associations related to socioeconomic status (household gross income and highest grade finished in school). For CHRNA3-CHRNA5 risk alleles, we replicated association with smoking behaviors, lung cancer, and asthma. There were also novel suggestive CHRNA3-CHRNA5 findings with respect to high-cholesterol-medication use and distrustful attitude. In conclusion, the genetics of alcohol and tobacco use potentially has broader implications on physical and mental health than is currently recognized. In particular, ADH1B may be a gene relevant for the human phenome via both alcohol metabolism-related mechanisms and other alcohol metabolism-independent mechanisms.
Figures
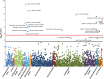
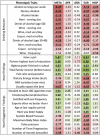


Similar articles
-
A phenome-wide association and Mendelian randomisation study of alcohol use variants in a diverse cohort comprising over 3 million individuals.EBioMedicine. 2024 May;103:105086. doi: 10.1016/j.ebiom.2024.105086. Epub 2024 Apr 4. EBioMedicine. 2024. PMID: 38580523 Free PMC article.
-
ADH1B: From alcoholism, natural selection, and cancer to the human phenome.Am J Med Genet B Neuropsychiatr Genet. 2018 Mar;177(2):113-125. doi: 10.1002/ajmg.b.32523. Epub 2017 Mar 27. Am J Med Genet B Neuropsychiatr Genet. 2018. PMID: 28349588 Free PMC article. Review.
-
Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study.Mol Psychiatry. 2017 Sep;22(9):1359-1367. doi: 10.1038/mp.2017.101. Epub 2017 May 9. Mol Psychiatry. 2017. PMID: 28485404 Free PMC article.
-
Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits.Nicotine Tob Res. 2012 Jun;14(6):720-33. doi: 10.1093/ntr/ntr283. Epub 2012 Jan 12. Nicotine Tob Res. 2012. PMID: 22241830 Free PMC article.
-
Biology, Genetics, and Environment: Underlying Factors Influencing Alcohol Metabolism.Alcohol Res. 2016;38(1):59-68. Alcohol Res. 2016. PMID: 27163368 Free PMC article. Review.
Cited by
-
Signatures of Convergent Evolution and Natural Selection at the Alcohol Dehydrogenase Gene Region are Correlated with Agriculture in Ethnically Diverse Africans.Mol Biol Evol. 2022 Oct 7;39(10):msac183. doi: 10.1093/molbev/msac183. Mol Biol Evol. 2022. PMID: 36026493 Free PMC article.
-
A genome-wide gene-by-trauma interaction study of alcohol misuse in two independent cohorts identifies PRKG1 as a risk locus.Mol Psychiatry. 2018 Jan;23(1):154-160. doi: 10.1038/mp.2017.24. Epub 2017 Mar 7. Mol Psychiatry. 2018. PMID: 28265120 Free PMC article.
-
The genetic epidemiology of substance use disorder: A review.Drug Alcohol Depend. 2017 Nov 1;180:241-259. doi: 10.1016/j.drugalcdep.2017.06.040. Epub 2017 Aug 1. Drug Alcohol Depend. 2017. PMID: 28938182 Free PMC article. Review.
-
Phenome-wide association study for CYP2A6 alleles: rs113288603 is associated with hearing loss symptoms in elderly smokers.Sci Rep. 2017 Apr 21;7(1):1034. doi: 10.1038/s41598-017-01098-4. Sci Rep. 2017. PMID: 28432340 Free PMC article.
-
A phenome-wide association and Mendelian randomisation study of alcohol use variants in a diverse cohort comprising over 3 million individuals.EBioMedicine. 2024 May;103:105086. doi: 10.1016/j.ebiom.2024.105086. Epub 2024 Apr 4. EBioMedicine. 2024. PMID: 38580523 Free PMC article.
References
-
- Buhler KM, Gine E, Echeverry-Alzate V, Calleja-Conde J, de Fonseca FR, Lopez-Moreno JA (2015). Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict Biol 20: 845–871. - PubMed
-
- Bush WS, Oetjens MT, Crawford DC (2016). Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 17: 129–145. - PubMed
MeSH terms
Substances
Grants and funding
- RC2 DA028909/DA/NIDA NIH HHS/United States
- N01 WH042111/WH/WHI NIH HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- N01 WH042107/WH/WHI NIH HHS/United States
- N01WH42109/WH/WHI NIH HHS/United States
- N01WH32111/WH/WHI NIH HHS/United States
- N01WH42117/WH/WHI NIH HHS/United States
- R01 AA017535/AA/NIAAA NIH HHS/United States
- N01WH42122/WH/WHI NIH HHS/United States
- P50 AA012870/AA/NIAAA NIH HHS/United States
- N01WH42119/WH/WHI NIH HHS/United States
- R01 DA012690/DA/NIDA NIH HHS/United States
- N01WH32112/WH/WHI NIH HHS/United States
- N01WH32101/WH/WHI NIH HHS/United States
- U01 HG004790/HG/NHGRI NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- R01 DA012849/DA/NIDA NIH HHS/United States
- N01WH32119/WH/WHI NIH HHS/United States
- N01WH42132/WH/WHI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- N01 WH042131/WH/WHI NIH HHS/United States
- N01WH42121/WH/WHI NIH HHS/United States
- R01 DA018432/DA/NIDA NIH HHS/United States
- N01 WH032102/WH/WHI NIH HHS/United States
- N01 WH042120/WH/WHI NIH HHS/United States
- N01WH42113/WH/WHI NIH HHS/United States
- N01WH42125/WH/WHI NIH HHS/United States
- N01 WH042130/WH/WHI NIH HHS/United States
- N01 WH042112/WH/WHI NIH HHS/United States
- N01WH32106/WH/WHI NIH HHS/United States
- N01WH42129/WH/WHI NIH HHS/United States
- N01 WH42124/OD/NIH HHS/United States
- N01WH42108/WH/WHI NIH HHS/United States
- N01 WH042114/WH/WHI NIH HHS/United States
- N01WH32113/WH/WHI NIH HHS/United States
- N01 WH032105/WH/WHI NIH HHS/United States
- N01 WH042126/WH/WHI NIH HHS/United States
- N01 WH042118/WH/WHI NIH HHS/United States
- N01WH32122/WH/WHI NIH HHS/United States
- R01 AA011330/AA/NIAAA NIH HHS/United States
- N01WH42115/WH/WHI NIH HHS/United States
- N01 WH032118/WH/WHI NIH HHS/United States
- N01WH32109/WH/WHI NIH HHS/United States
- N01WH32108/WH/WHI NIH HHS/United States
- N01WH32100/WH/WHI NIH HHS/United States
- N01WH42123/WH/WHI NIH HHS/United States
- N01WH32115/WH/WHI NIH HHS/United States
- N01 WH042116/WH/WHI NIH HHS/United States
- N01WH44221/WH/WHI NIH HHS/United States
- N01WH24152/WH/WHI NIH HHS/United States
- U01 HG004801/HG/NHGRI NIH HHS/United States
- N01WH42110/WH/WHI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

