A Novel Method to Verify Multilevel Computational Models of Biological Systems Using Multiscale Spatio-Temporal Meta Model Checking
- PMID: 27187178
- PMCID: PMC4871515
- DOI: 10.1371/journal.pone.0154847
A Novel Method to Verify Multilevel Computational Models of Biological Systems Using Multiscale Spatio-Temporal Meta Model Checking
Abstract
Insights gained from multilevel computational models of biological systems can be translated into real-life applications only if the model correctness has been verified first. One of the most frequently employed in silico techniques for computational model verification is model checking. Traditional model checking approaches only consider the evolution of numeric values, such as concentrations, over time and are appropriate for computational models of small scale systems (e.g. intracellular networks). However for gaining a systems level understanding of how biological organisms function it is essential to consider more complex large scale biological systems (e.g. organs). Verifying computational models of such systems requires capturing both how numeric values and properties of (emergent) spatial structures (e.g. area of multicellular population) change over time and across multiple levels of organization, which are not considered by existing model checking approaches. To address this limitation we have developed a novel approximate probabilistic multiscale spatio-temporal meta model checking methodology for verifying multilevel computational models relative to specifications describing the desired/expected system behaviour. The methodology is generic and supports computational models encoded using various high-level modelling formalisms because it is defined relative to time series data and not the models used to generate it. In addition, the methodology can be automatically adapted to case study specific types of spatial structures and properties using the spatio-temporal meta model checking concept. To automate the computational model verification process we have implemented the model checking approach in the software tool Mule (http://mule.modelchecking.org). Its applicability is illustrated against four systems biology computational models previously published in the literature encoding the rat cardiovascular system dynamics, the uterine contractions of labour, the Xenopus laevis cell cycle and the acute inflammation of the gut and lung. Our methodology and software will enable computational biologists to efficiently develop reliable multilevel computational models of biological systems.
Conflict of interest statement
Figures

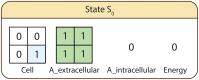




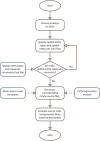


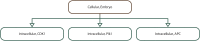


Similar articles
-
Verifiable biology.J R Soc Interface. 2023 May;20(202):20230019. doi: 10.1098/rsif.2023.0019. Epub 2023 May 10. J R Soc Interface. 2023. PMID: 37160165 Free PMC article. Review.
-
Automatic validation of computational models using pseudo-3D spatio-temporal model checking.BMC Syst Biol. 2014 Dec 2;8:124. doi: 10.1186/s12918-014-0124-0. BMC Syst Biol. 2014. PMID: 25440773 Free PMC article.
-
Automatic selection of verification tools for efficient analysis of biochemical models.Bioinformatics. 2018 Sep 15;34(18):3187-3195. doi: 10.1093/bioinformatics/bty282. Bioinformatics. 2018. PMID: 29688313 Free PMC article.
-
STSE: Spatio-Temporal Simulation Environment Dedicated to Biology.BMC Bioinformatics. 2011 Apr 28;12:126. doi: 10.1186/1471-2105-12-126. BMC Bioinformatics. 2011. PMID: 21527030 Free PMC article.
-
Computational Modeling, Formal Analysis, and Tools for Systems Biology.PLoS Comput Biol. 2016 Jan 21;12(1):e1004591. doi: 10.1371/journal.pcbi.1004591. eCollection 2016 Jan. PLoS Comput Biol. 2016. PMID: 26795950 Free PMC article. Review.
Cited by
-
Verifiable biology.J R Soc Interface. 2023 May;20(202):20230019. doi: 10.1098/rsif.2023.0019. Epub 2023 May 10. J R Soc Interface. 2023. PMID: 37160165 Free PMC article. Review.
-
Formal reasoning about systems biology using theorem proving.PLoS One. 2017 Jul 3;12(7):e0180179. doi: 10.1371/journal.pone.0180179. eCollection 2017. PLoS One. 2017. PMID: 28671950 Free PMC article.
-
Systems Medicine-Complexity Within, Simplicity Without.J Healthc Inform Res. 2017;1(1):119-137. doi: 10.1007/s41666-017-0002-9. Epub 2017 May 10. J Healthc Inform Res. 2017. PMID: 28713872 Free PMC article.
-
An Integrative Analysis of Time-varying Regulatory Networks From High-dimensional Data.Proc IEEE Int Conf Big Data. 2018 Dec;2018:3798-3807. doi: 10.1109/BigData.2018.8622361. Epub 2019 Jan 24. Proc IEEE Int Conf Big Data. 2018. PMID: 31544173 Free PMC article.
-
Coloured Petri nets for multilevel, multiscale and multidimensional modelling of biological systems.Brief Bioinform. 2019 May 21;20(3):877-886. doi: 10.1093/bib/bbx150. Brief Bioinform. 2019. PMID: 29112705 Free PMC article. Review.
References
-
- Ideker T, Galitski T, Hood L. A NEW APPROACH TO DECODING LIFE: Systems Biology. Annual Review of Genomics and Human Genetics. 2001;2(1):343–372. Available from: http://www.annualreviews.org/doi/abs/10.1146/annurev.genom.2.1.343. 10.1146/annurev.genom.2.1.343 - DOI - DOI - PubMed
-
- Kitano H. Systems Biology: A Brief Overview. Science. 2002. January;295(5560):1662–1664. Available from: http://www.sciencemag.org/content/295/5560/1662. 10.1126/science.1069492 - DOI - PubMed
-
- Boissel JP, Auffray C, Noble D, Hood L, Boissel FH. Bridging Systems Medicine and Patient Needs. CPT: Pharmacometrics & Systems Pharmacology. 2015. March;4(3):135–145. Available from: http://onlinelibrary.wiley.com/doi/10.1002/psp4.26/abstract. - DOI - PMC - PubMed
-
- Wolkenhauer O, Auffray C, Brass O, Clairambault J, Deutsch A, Drasdo D, et al. Enabling multiscale modeling in systems medicine. Genome Medicine. 2014. March;6(3):21 Available from: http://genomemedicine.com/content/6/3/21. 10.1186/gm538 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

