A two-part mixed-effects model for analyzing longitudinal microbiome compositional data
- PMID: 27187200
- PMCID: PMC5860434
- DOI: 10.1093/bioinformatics/btw308
A two-part mixed-effects model for analyzing longitudinal microbiome compositional data
Abstract
Motivation: The human microbial communities are associated with many human diseases such as obesity, diabetes and inflammatory bowel disease. High-throughput sequencing technology has been widely used to quantify the microbial composition in order to understand its impacts on human health. Longitudinal measurements of microbial communities are commonly obtained in many microbiome studies. A key question in such microbiome studies is to identify the microbes that are associated with clinical outcomes or environmental factors. However, microbiome compositional data are highly skewed, bounded in [0,1), and often sparse with many zeros. In addition, the observations from repeated measures in longitudinal studies are correlated. A method that takes into account these features is needed for association analysis in longitudinal microbiome data.
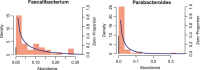
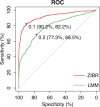
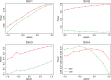
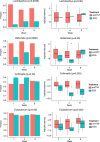
Results: In this paper, we propose a two-part zero-inflated Beta regression model with random effects (ZIBR) for testing the association between microbial abundance and clinical covariates for longitudinal microbiome data. The model includes a logistic regression component to model presence/absence of a microbe in the samples and a Beta regression component to model non-zero microbial abundance, where each component includes a random effect to account for the correlations among the repeated measurements on the same subject. Both simulation studies and the application to real microbiome data have shown that ZIBR model outperformed the previously used methods. The method provides a useful tool for identifying the relevant taxa based on longitudinal or repeated measures in microbiome research.
Availability and implementation: https://github.com/chvlyl/ZIBR CONTACT: hongzhe@upenn.edu.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Arrieta M.C. et al. . ( 2015. ) Early infancy microbial and metabolic alterations affect risk of childhood asthma . Sci. Transl. Med ., 7 , 307ra152 – 307ra152 . - PubMed
-
- Bäckhed F. et al. . ( 2015. ) Dynamics and stabilization of the human gut microbiome during the first year of life . Cell Host Microbe , 17 , 690 – 703 . - PubMed
-
- Benjamini Y., Hochberg Y. ( 1995. ) Controlling the false discovery rate: a practical and powerful approach to multiple testing . J. R. Stat. Soc. Ser. B , 57 , 289 – 300 .
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

