Chemical Variations on the p53 Reactivation Theme
- PMID: 27187415
- PMCID: PMC4932543
- DOI: 10.3390/ph9020025
Chemical Variations on the p53 Reactivation Theme
Abstract
Among the tumor suppressor genes, p53 is one of the most studied. It is widely regarded as the "guardian of the genome", playing a major role in carcinogenesis. In fact, direct inactivation of the TP53 gene occurs in more than 50% of malignancies, and in tumors that retain wild-type p53 status, its function is usually inactivated by overexpression of negative regulators (e.g., MDM2 and MDMX). Hence, restoring p53 function in cancer cells represents a valuable anticancer approach. In this review, we will present an updated overview of the most relevant small molecules developed to restore p53 function in cancer cells through inhibition of the p53-MDMs interaction, or direct targeting of wild-type p53 or mutated p53. In addition, optimization approaches used for the development of small molecules that have entered clinical trials will be presented.
Keywords: MDM2 inhibitors; MDMX inhibitors; mutant p53; p53 activators; p53-MDM2 interaction inhibitors; p53-MDMX interaction inhibitors; small molecules; wild-type p53.
Figures
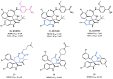
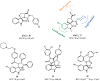
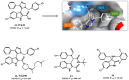
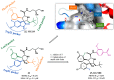
Similar articles
-
An Update on MDMX and Dual MDM2/X Inhibitors.Curr Top Med Chem. 2018;18(8):647-660. doi: 10.2174/1568026618666180604080119. Curr Top Med Chem. 2018. PMID: 29866007 Review.
-
1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.J Med Chem. 2017 May 25;60(10):4234-4244. doi: 10.1021/acs.jmedchem.7b00104. Epub 2017 May 16. J Med Chem. 2017. PMID: 28482147
-
MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer.J Hematol Oncol. 2017 Jul 3;10(1):133. doi: 10.1186/s13045-017-0500-5. J Hematol Oncol. 2017. PMID: 28673313 Free PMC article. Review.
-
Medicinal Chemistry Strategies to Disrupt the p53-MDM2/MDMX Interaction.Med Res Rev. 2016 Sep;36(5):789-844. doi: 10.1002/med.21393. Epub 2016 Jun 15. Med Res Rev. 2016. PMID: 27302609 Review.
-
A tryptophanol-derived oxazolopiperidone lactam is cytotoxic against tumors via inhibition of p53 interaction with murine double minute proteins.Pharmacol Res. 2015 May-Jun;95-96:42-52. doi: 10.1016/j.phrs.2015.03.006. Epub 2015 Mar 23. Pharmacol Res. 2015. PMID: 25814188
Cited by
-
Molecular Biology of Osteosarcoma.Cancers (Basel). 2020 Jul 31;12(8):2130. doi: 10.3390/cancers12082130. Cancers (Basel). 2020. PMID: 32751922 Free PMC article. Review.
-
Recent Synthetic Approaches towards Small Molecule Reactivators of p53.Biomolecules. 2020 Apr 20;10(4):635. doi: 10.3390/biom10040635. Biomolecules. 2020. PMID: 32326087 Free PMC article. Review.
-
Regio- and stereoselective synthesis of spiropyrrolidine-oxindole and bis-spiropyrrolizidine-oxindole grafted macrocycles through [3 + 2] cycloaddition of azomethine ylides.RSC Adv. 2020 Mar 10;10(17):10263-10276. doi: 10.1039/c9ra10463a. eCollection 2020 Mar 6. RSC Adv. 2020. PMID: 35498613 Free PMC article.
-
A Novel Small Molecule p53 Stabilizer for Brain Cell Differentiation.Front Chem. 2019 Jan 31;7:15. doi: 10.3389/fchem.2019.00015. eCollection 2019. Front Chem. 2019. PMID: 30766866 Free PMC article.
-
The Double Role of p53 in Cancer and Autoimmunity and Its Potential as Therapeutic Target.Int J Mol Sci. 2016 Nov 25;17(12):1975. doi: 10.3390/ijms17121975. Int J Mol Sci. 2016. PMID: 27897991 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

